Abstract
Inflammatory bowel disease (IBD) is a chronic and heterogeneous disorder characterized by remitting and relapsing periods of activity. Pharmacogenetics refers to the study of the effect of inheritance on individual variation in drug responses. Several drug-related markers in IBD patients have been identified in order to predict the response to medical treatment including biological therapy as well as the reduction of adverse events. In the future, the treatment of IBD should be personalized in its specific profile to provide the most efficacious treatment with lack of adverse events.
Introduction
Inflammatory bowel disease (IBD) comprises Crohn’s disease (CD) and ulcerative colitis (UC) characterized by intestinal chronic inflammation of unknown etiology. It has been postulated that it is a multifactorial disease involving interplay among aberrant immune response, environmental factors, and multiple genes.Citation1
The incidence of IBD is now rising in developing countries, and it is being increasingly considered an emerging global disease.Citation2,Citation3 Traditionally, developing nations have reported a lower prevalence of IBD, but the incidence is currently rising in many of these countries as they become more industrialized in Latin America and Asia.Citation4–Citation7
Knowledge development in IBD has permitted classification of the disease into different phenotypes according to the current clinical Montreal classification that considers clinical characteristics such as age at diagnosis, location of disease, behavior in CD, and extent of disease in UC.Citation8 Clinical traits such as the extent of inflammation or disease location, extraintestinal manifestations, and disease behavior enabled predicting the future course of the disease and response to medical therapy to allow disease categorization which in turn guides present-day care recommendations.Citation9
On the other hand, understanding the involvement of several molecular pathways by different technologies such as genomic, transcriptomic, epigenetic, and miRNAs studies has been focused on the identification of new genetic risk factors for specific disease and application to clinical practice such as prognostic factors involved in the clinical course and response to medical therapy.Citation10,Citation11
Finally, pharmacogenetics is the study of the association between variability in drug response, drug toxicity, and polymorphisms in genes in order to adapt drugs to a patient’s specific genetic background and therefore make them more efficacious and safe.Citation12
The vast heterogeneity of IBD patients has motivated a comprehensive evidence-based search of novel biomarkers for appropriate patient stratification that accounts for the interindividual differences in severity, drug efficacy, side effects, or prognosis and would help guide clinicians in the management of patients with IBD and represents a major step toward personalized medicine.Citation13
This study aimed to review the role of pharmacogenetics in the potential selection of personalized treatment in patients with IBD in the future.
Several drug-related markers classified according to pharmacological groups with clinical utility in patients with IBD are described below and summarized in .
Table 1 Drug-related markers in inflammatory bowel disease
Pharmacogenetics in IBD treatment
Mesalazine (5-aminosalicylic acid [5-ASA])
Mesalazine constitutes the first line of treatment for induction and maintenance of remission in UC. It appears to act locally on colonic mucosa and reduces inflammation through a variety of anti-inflammatory processes by the activation of γ-form peroxisome proliferator-activated receptors (PPAR-γ). PPAR-γ has a role in the regulation of intestinal inflammation and is highly expressed in the colon, where epithelial cells and macrophages are the main cellular sources of this nuclear receptor.Citation14
A study showed decreased gene expression of PPAR-γ in colonic biopsies of patients with active UC and its expression was negatively correlated with severity of endoscopic disease activity.Citation15
In another study, the PPAR alpha gene expression was significantly decreased in patients with active UC compared with remission UC group (P=0.001) and normal controls (P=0.001). Yamamoto-Furusho et alCitation16 found that low gene expression of PPAR alpha in colonic mucosa is associated with high risk of UC activity (P≤0.0001, odds ratio [OR]=22.6). They observed an increase of PPAR alpha expression in patients with UC who were treated with 5-aminosalicylates compared with those who received any other combined therapy (P=0.03, OR=0.08). PPAR-γ gene expression was decreased in the active UC group compared with remission UC (P=0.001) and control group (P=0.001). An increased expression of PPAR-γ gene was associated with mild clinical course of the disease (P≤0.001, OR=0.05).Citation16
Heat shock proteins (Hsp) are a family of molecules typically involved in folding, refolding, translocation, and degradation of intracellular proteins under normal and stress conditions.Citation17 Hsp60 and Hsp10 (Hsp60 co-chaperonin) are increased in the affected intestinal mucosa in patients with CD and UC.Citation18
A study demonstrated that mucosal Hsp60 levels in UC patients decrease after therapy with either mesalazine alone or mesalazine plus probiotics. They also demonstrated that Hsp90 levels are elevated in colonic mucosa from UC patients, both in epithelium and lamina propria.Citation19 Treatment with 5-ASA plus probiotics reduced the Hsp90 levels in the lamina propria, while 5-ASA alone did not have any effect and a linear correlation was also reported between Hsp90 and CD4 levels in lamina propria in UC patients at both diagnosis and 6 months after 5-ASA only therapy.Citation20
Corticosteroids
Corticosteroids constitute the second line of therapy in patients who fail to respond to the maximal dose of mesalazine or present moderate-to-severe disease activity of IBD. The mechanism of action of corticosteroids is based on the inhibition of T-cell activation and the production of proinflammatory cytokines.
A review has proposed different markers associated with steroid therapy outcomes in patients with IBD.Citation21 The most-studied molecule is the multidrug-resistant (MDR1) gene code for a drug efflux pump P-glycoprotein-170 (permeability glycoprotein or Pgp), which is expressed on the apical surface of lymphocytes and intestinal epithelial cells. Its function comprises active transportation of toxins and drugs out of target cells.Citation22 Pgp and MDR expression have been shown to be significantly higher in CD and UC patients requiring surgery due to failure of medical therapy.Citation23 On the other hand, MDR1 expression on colonic biopsies decreased in patients with active UC compared to UC patients in remission and the normal control group (P=0.034 and 0.002, respectively). However, in this study, the relevant finding was that the medical treatment response and long-term remission were associated with high gene expression of MDR1 (P=0.009 and 0.002, respectively).Citation24
Other studies have reported an increased expression of glucocorticoid receptor β in 83% of the patients with steroid-resistant UC compared to only 9% in steroid-responsive patients and 10% in healthy controls.Citation25,Citation26
Yamamoto-Furusho et alCitation27 found a significant association between RN6/2 GG (rs315951) and interleukin (IL)-1B-511 CC (rs16944) genotypes and the presence of steroid dependence in UC patients (p corrected=0.0001, OR=15.6 and pC=0.008, OR=4.09, respectively).
A previous study identified several predictor gene panels containing genes involved in immune mechanisms (PTN, OLFM4, LILRA2, CD36), autophagy, or GC response (STS, MDM2) with potential value to predict GC response and need of surgery as well as with diagnostic value for IBD patients.Citation28 Villeda-Ramirez et alCitation29 showed that IL-18 mRNA expression was significantly increased in the mucosa of patients with active and remission UC compared to the healthy control group (P=0.006 and 0.007, respectively). The high gene expression of IL-18 was associated with the use of steroids (P=0.04).
Thiopurines
Immunomodulator drugs have become the mainstay of IBD with proven efficacy in reducing relapses, permitting steroid withdrawal, and closing fistulas.Citation30
Thiopurines such as azathioprine (AZA) and 6-mercaptopurine (6-MP) are usually used in patients with corticosteroid dependence or resistance and combined with tumor necrosis factor (TNF) therapy. The gene encoding thiopurine methyltransferase (TPMT) is located on chromosome 6 (6p22.3) and contains 10 exons. Two wild-type alleles (TPMT*1 and *1S) and 20 mutant alleles (TPMT*2, *3A, *3B, *3C, *3D, *4, *5, *6, *7, *8, *9, *10, *11,*12, *13, *14, *15, *16, *17, *18) are responsible for TPMT deficiency.Citation31,Citation32 The distribution of TPMT mutant alleles differs significantly among ethnic populations. TPMT*3A (3.2%–5.7%) is the most occurring mutant allele in white populations, followed by TPMT*2 (0.2%–0.5%) and TPMT*3C (0.2%–0.8%), accounting for the vast majority (>95%) of mutant alleles.Citation33–Citation36 In Asian and African populations, however, TPMT*3C is the most frequent mutant allele.Citation37,Citation38
A genome-wide association study found a 2.59-fold risk of pancreatitis in IBD patients taking thiopurines who had the single nucleotide polymorphism rs2647087 within the human leukocyte antigen (HLA)-DQA1*02:01-HLA-DRB1*07:01 haplotype.Citation39
Traditionally, AZA and 6-MP are initiated at a low dose and then gradually increased to a full therapeutic dose of 2.0–2.5 and 1.5 mg/kg/day, respectively; however, this strategy requires tight monitoring in order to detect adverse events such as myelotoxicity and hepatotoxicity. This strategy has been replaced by an approach based on the assessment of TPMT phenotype or activity as shown in . TPMT testing is recommended before initiating AZA or 6-MP therapy for IBD to decrease the risk of leukopenia. For patients who have absent or low TPMT, activity leading to elevated 6-thioguanine nucleotide (6-TGN) concentrations during thiopurine therapy is significantly associated with an increased risk of development of bone marrow suppression.Citation40 In patients with very high TPMT, activity develops suboptimal 6-TGN concentrations, which have been associated with treatment failure.Citation41 Population studies have shown that the distribution of TPMT activity is trimodal: 0.3% of the population have low-to-absent activity, 11% have intermediate activity, and 89% inherit normal-to-high enzyme activity.Citation42–Citation44
A review by Stocco et alCitation45 has reported the role of enzyme glutathione-S-transferase (GST) genetic polymorphisms which may influence decreased sensitivity to AZA. Some studies have found that the frequency of GST-M1 deletion was significantly lower in patients who developed an adverse event in comparison to patients who tolerated AZA treatment with no adverse event.Citation46–Citation48
On the other hand, measurement of the thiopurine metabolites, 6-TGN and 6-methylmercaptopurine (6-MMP), is useful in clinical practice. Several studies have demonstrated that 6-TGN levels >230 pmol/8×108 red blood cells (RBCs) are associated with increased efficacy.Citation49,Citation50 However, supra-therapeutic levels, generally >400 pmol/8×108 RBCs, are associated with an increased risk of myelosuppression.Citation51 6-MMP can be measured to predict the risk of hepatotoxicity; levels >5,700 pmol/8×108 RBCs carry a 3-fold risk of hepatotoxicity.Citation52
Anti-TNF therapy
Anti-TNF-α drugs are indicated in patients with moderate-to-severe IBD who do not tolerate or respond to conventional therapies. The use of anti-TNF therapy has improved several outcomes in patients with IBD such as better quality of life, reduction of surgeries and hospitalizations, steroid free remission, mucosal healing, and others. However, one third of the patients do not respond to anti-TNF treatment.
Several studies have focused on studying genetic markers that may predict individual response to anti-TNF therapy.Citation53 In luminal CD, the response rate to anti-TNF therapy was 74.7% in patients with Fas ligand (FASLG) −843 CC/CT genotype compared to a response rate of 38.1% in patients with the TT genotype (OR=0.11; 95% confidence interval [CI]=0.08–0.56, P<0.01). On the other hand, patients with caspase-9 TT genotype responded to anti-TNF therapy, in contrast to 66.7% of patients with the CC and CT genotypes (OR=1.50; 95% CI=1.34–1.68, P=0.04). Another variant in FASLG, rs763110, was able to predict the therapeutic response to infliximab in patients with fistulizing CD at week 10.Citation54 A Japanese study reported that GG genotype of FCGR3A had a better response to anti-TNF therapy at week 8 in CD patients.Citation55
Autophagy-related 16-like 1 (ATG16L1) is an autophagy-related gene that processes intracellular bacteria and is expressed in intestinal epithelial cell lines. ATG16L1 TT genotype for rs10210302 responded better to adalimumab after 12, 20, and 30 weeks of treatment compared to the CC genotype in CD patients.Citation56
The cytokine IL-23 is involved in the pathogenesis of IBD. Several genetic variants in IL-23R have been associated with response to infliximab in patients with moderate-to-severe UC at week 14. For instance, AA genotype for rs1004819, rs10889677, and rs11209032, GG genotype for rs2201841, and CC genotype for rs1495965 in IL-23R gene increased the probability to respond to infliximab. However, GG genotype for rs7517847 and rs11465804, CC genotype for rs10489629, and AA genotype for rs1343151 in IL-23R decreased the probability to respond to this drug.Citation57 Bank et al found that the TC/CC genotype for rs10499563 in IL-6 and the GA/AA genotype had a better response to anti-TNF but the effect of IL-1RN (rs4251961) allele C was associated with poorer response to anti-TNF therapy.Citation58
In some patients who had a genetically increased MD-2 level (rs11465996) and TNFRSF1A (TNFR1) expression (rs4149570) and genetically determined decreased TNFAIP3 (A20) expression (rs6927172), IL-1β (rs3804099 and rs4848306), IL-6 (rs3804099 and rs10499563), IL-17 (rs2275913), and interferon-γ (rs2430561) levels were associated with beneficial response among patients with IBD. Fujino et alCitation59 found mRNA expression and serum levels of IL-17 to be increased in patients with IBD and suggested that IL-17 might be associated with altered immune and inflammatory responses in the intestinal mucosa.
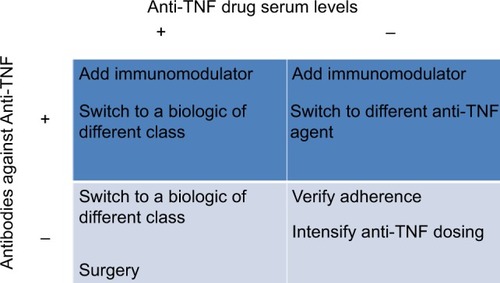
Therapeutic drug monitoring (TDM) and measurement of antidrug antibodies (ADAs) for anti-TNF agents have been useful in clinical practice to optimize the efficacy of biologics and minimize adverse events (). TDM has been best studied for infliximab and adalimumab including the measurement of both drug and antibodies to infliximab (ATIs) or antibodies to adalimumab (ATAs). Several studies have reported concentrations predictive of response ranging from 1.4 to 12.0 μg/mL.Citation60–Citation63 For adalimumab, cutoffs predictive of mucosal healing range from 4.9 to 7.5 μg/mL.Citation64,Citation65 Low levels of anti-TNF agents are associated with developing ADAs and preceded the formation of ATIs and ATAsCitation66,Citation67 and these are implicated in the increased drug clearance of anti-TNF agents related with lower serum drug levels as well as active disease and loss of response.Citation68,Citation69
Anti-integrin therapy
Etrolizumab is a monoclonal antibody against β7 integrin subunit that has shown efficacy in patients with moderate-to-severe UC. A study evaluated biomarkers in the colonic mucosa such as granzyme A and integrin αE measured by gene and protein expression levels; both markers were found to be associated with clinical response and mucosal healing in UC patients treated with etrolizumab.Citation70
Microbiota
A potential link between genetics and the microbiome has been documented in patients with IBD. Some studies have shown the effect of NOD2 mutations associated with increased numbers of mucosa-adherent bacteriaCitation71 and decreased transcription of the anti-inflammatory cytokine IL-10.Citation72
IBD patients with NOD2 and ATG16L1 have significant alterations in the structure of their gut microbiota, including decreased levels of Faecalibacterium and increases in Escherichia.Citation73 Individuals homozygous for loss-of-function alleles for fucosyltransferase 2 are “nonsecretors,” who do not express ABO antigen on the gastrointestinal mucosa and bodily secretions. Nonsecretors are at increased risk for CD28 and exhibit substantial alterations in the mucosa-associated microbiota.Citation74
Several studies have shown specific taxonomic shifts in IBD patients. Enterobacteriaceae are increased in IBD patients.Citation75 Escherichia coli, particularly adherent-invasive E. coli strains, has been isolated from CD biopsies in culture-based studiesCitation76 and is enriched in UC patients.Citation77 Another type of adherent and invasive bacteria is Fusobacteria. The genus Fusobacterium has been found in higher abundance in the colonic mucosa of patients with UC relative to control individuals.Citation78,Citation79
On the other hand, specific groups of gut bacteria may have protective effects against IBD. Bacteroides and Clostridium species have been shown to induce the expansion of Treg cells, reducing intestinal inflammation.Citation80
Other bacterial species such as Bifidobacterium, Lactobacillus, and Faecalibacterium may protect the host from mucosal inflammation by several mechanisms, including downregulation of inflammatory cytokinesCitation81 or stimulation of IL-10.Citation82 Faecalibacterium prausnitzii is a member of the microbiota with anti-inflammatory properties. F. prausnitzii has been found depleted in CD biopsy samples concomitant with an increase in E. coli abundance,Citation83 and low levels of mucosa-associated F. prausnitzii are associated with higher risk of recurrent CD following surgery.Citation82 Conversely, recovery of F. prausnitzii after relapse is associated with maintenance of clinical remission of UC.Citation84
A recent study has shown a microbial signature for CD that identified eight groups of microorganisms including Faecalibacterium, Peptostreptococcaceae, Anaerostipes, Methanobrevibacter, Christensenellaceae, Collinsella, Fusobacterium, and Escherichia; the signature achieved an overall sensitivity of 80% and a specificity of 94% for the detection of CD versus healthy controls.Citation85
Fecal microbiota transplantation (FMT) aims to restore gut microbiota diversity by transferring feces from a healthy donor to a sick patient. To date, FMT has been assessed as a novel therapeutic for UC. A randomized controlled trial reported 24% and 5% of those who received FMT and placebo, respectively, reached clinical remission after 7 weeks. This study identified that those patients who had been recently diagnosed of UC (within the year previous to FMT) had a high rate of clinical remission compared to UC patients with longer disease duration (75% vs 18% respectively).Citation86
On the other hand, another study by Rossen et alCitation87 showed no statistically significant difference in clinical remission between UC patients who received FMT sourced from healthy donors or autologous FMT (their own fecal microbiota), and only 41% of patients who received donor FMT achieved clinical and endoscopic remission. The findings of this study suggest that microbial ecosystems of patients who responded to FMT from a healthy donor increased in the numbers of bacterial species from Clostridium clusters.
A detailed assessment of the fecal microbiota taxonomic composition pre- and post-FMT need to be performed in order to identify the responders to FMT in patients with UC. A selective FMT of certain species such as Bifidobacterium, Lactobacillus, and F. prausnitzii could be effective as personalized treatment in patients with IBD.
In conclusion, the combination of genetic markers with clinical, biochemical, serological, and microbiome data for subgroups of IBD patients might permit individualized risk stratification and treatment selection to ensure high efficacy of medical treatment with lack of adverse events.
Disclosure
The author has received honoraria from AbbVie, Takeda, Janssen, UCB, Almirall, Pfizer, Novartis, and Danone as speaker, key opinion leader, and member of the advisory board at national and international levels. He is the President of the Pan American Crohn’s and Colitis Organization.
References
- Yamamoto-FurushoJKPodolskyDKInnate immunity in inflammatory bowel diseaseWorld J Gastroenterol2007135577558017948931
- BernsteinCNShanahanFDisorders of a modern lifestyle: reconciling the epidemiology of inflammatory bowel diseasesGut20085791185119118515412
- GohKXiaoSDInflammatory bowel disease: a survey of the epidemiology in AsiaJ Dig Dis2009101619236540
- TorresEADe JesúsRPérezCMPrevalence of inflammatory bowel disease in an insured population in Puerto Rico during 1996P R Health Sci J20032225325814619451
- Yamamoto-FurushoJKClinical epidemiology of ulcerative colitis in Mexico: a single hospital-based study in a 20-year period (1987–2006)J Clin Gastroenterol20094322122419057395
- NgSCTangWChingJYIncidence and phenotype of inflammatory bowel disease based on results from the Asia-pacific Crohn’s and colitis epidemiology studyGastroenterology201314515816523583432
- ParenteJMCoyCSCampeloVInflammatory bowel disease in an underdeveloped region of Northeastern BrazilWorld J Gastroenterol2015211197120625632193
- SilverbergMSSatsangiJAhmadTToward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a Working Party of the 2005 Montreal World Congress of GastroenterologyCan J Gastroenterol200519Suppl A5A36A
- GerichMEMcGovernDPTowards personalized care in IBDNat Rev Gastroenterol Hepatol20141128729924345887
- Yamamoto-FurushoJKFonseca-CamarilloGGenetic markers associated with clinical outcomes in patients with inflammatory bowel diseaseInflamm Bowel Dis2015212683269526244649
- QuetglasEGMujagicZWiggeSUpdate on pathogenesis and predictors of response of therapeutic strategies used in inflammatory bowel diseaseWorld J Gastroenterol201521125191254326640330
- KirchheinerJFuhrUBrockmöllerJPharmacogenetics-based therapeutic recommendations – ready for clinical practice?Nat Rev Drug Discov2005463964716056390
- KatsanosKHPapadakisKAPharmacogenetics of inflammatory bowel diseasePharmacogenomics2014152049206225521361
- LawsonMMThomasAGAkobengAKTumour necrosis factor alpha blocking agents for induction of remission in ulcerative colitis [review]Cochrane Database Syst Rev20063CD005112
- Yamamoto-FurushoJKPeñaloza-CoronelASánchez-MuñozFBarreto-ZúñigaRDomínguez-LópezAPeroxisome proliferator-activated receptor-gamma (PPAR gamma) expression is downregulated in patients with active ulcerative colitisInflamm Bowel Dis20111768068120848495
- Yamamoto-FurushoJKJacintez-CazaresMFuruzawa-CarballedaJFonseca-CamarilloGPeroxisome proliferator-activated receptors family is involved in the response to treatment and mild clinical course in patients with ulcerative colitisDis Markers2014201493253025548431
- ProhászkaZFüstGImmunological aspects of heat-shock proteins – the optimum stress of lifeMol Immunol200441294415140573
- RodolicoVTomaselloGZerilliMHsp60 and Hsp10 increase in colon mucosa of Crohn’s disease and ulcerative colitisCell Stress Chaperones20101587788420390473
- TomaselloGRodolicoVZerilliMChanges in immunohistochemical levels and subcellular localization after therapy and correlation and colocalization with CD68 suggest a pathogenetic role of Hsp60 in ulcerative colitisAppl Immunohistochem Mol Morphol20111955256121441812
- TomaselloGSciuméCRappaFHsp10, Hsp70, and Hsp90 immunohistochemical levels change in ulcerative colitis after therapyEur J Histochem201155e3822297444
- GabryelMSkrzypczak-ZielinskaMKucharskiMASlomskiRDobrowolskaAThe impact of genetic factors on response to glucocorticoids therapy in IBDScand J Gastroenterol20165165466526776488
- AnneseVValvanoMRPalmieriOMultidrug resistance 1 gene in inflammatory bowel disease: a meta-analysisWorld J Gastroenterol2006123636364416773678
- FarrellRJMurphyALongAHigh multidrug resistance (P-glycoprotein 170) expression in inflammatory bowel disease patients who fail medical therapyGastroenterology200011827928810648456
- Yamamoto-FurushoJKVilleda-RamírezMAFonseca-CamarilloGHigh gene expression of MDR1 (ABCB1) is associated with medical treatment response and long term remission in patients with ulcerative colitisInflamm Bowel Dis20101654171919714749
- HondaMOriiFAyabeTExpression of glucocorticoid receptor beta in lymphocytes of patients with glucocorticoid-resistant ulcerative colitisGastroenterology200011885986610784585
- FujishimaSTakedaHKawataSYamakawaMThe relationship between the expression of the glucocorticoid receptor in biopsied colonic mucosa and the glucocorticoid responsiveness of ulcerative colitis patientsClin Immunol200913320821719646928
- Yamamoto-FurushoJKSantiago-HernándezJJPérez-HernándezNRamírez-FuentesSFragosoJMVargas-AlarcónGInterleukin 1β (IL-1B) and IL-1 antagonist receptor (IL-1RN) gene polymorphisms are associated with the genetic susceptibility and steroid dependence in patients with ulcerative colitisJ Clin Gastroenterol20114553153520975573
- Montero-MeléndezTLlorXGarcía-PlanellaEPerrettiMSuárezAIdentification of novel predictor classifiers for inflammatory bowel disease by gene expression profilingPLoS One20138e7623524155895
- Villeda RamírezMAMendivil RangelEJDomínguez LópezAYamamoto-FurushoJKInterleukin-18 upregulation is associated with the use of steroids in patients with ulcerative colitisInflamm Bowel Dis201117E50E5121456035
- DubinskyMCAzathioprine, 6-mercaptopurine in inflammatory bowel disease: pharmacology, efficacy, and safetyClin Gastroenterol Hepatol2004273174315354273
- SchaeffelerELangTZangerUMEichelbaumMSchwabMHigh-throughput genotyping of thiopurine S-methyltransferase by denaturing HPLCClin Chem20014754855511238310
- SchaeffelerEFischerCBrockmeierDComprehensive analysis of thiopurine S-methyltransferase phenotype–genotype correlation in a large population of German-Caucasians and identification of novel TPMT variantsPharmacogenetics20041440741715226673
- ReutherLOSonneJLarsenNDahlerupJFThomsenOOSchmiegelowKThiopurine methyltransferase genotype distribution in patients with Crohn’s diseaseAliment Pharmacol Ther200317656812492733
- YatesCRKrynetskiEYLoennechenTMolecular diagnosis of thiopurine S-methyltransferase deficiency: genetic basis for azathioprine and mercaptopurine intoleranceAnn Intern Med19971266086149103127
- AnsariAHassanCDuleyJThiopurine methyltransferase activity and the use of azathioprine in inflammatory bowel diseaseAliment Pharmacol Ther2002161743175012269967
- KurzawskiMGawronska-SzklarzBDrozdzikMFrequency distribution of thiopurine S-methyltransferase alleles in a Polish populationTher Drug Monit20042654154515385838
- KubotaTChibaKFrequencies of thiopurine S-methyltransferase mutant alleles (TPMT*2, *3A, *3B and *3C) in 151 healthy Japanese subjects and the inheritance of TPMT*3C in the family of a propositusBr J Clin Pharmacol20015147547711422006
- AmeyawMMCollie-DuguidESPowrieRHOfori-AdjeiDMcLeodHLThiopurine methyltransferase alleles in British and Ghanaian populationsHum Mol Genet199983673709931345
- HeapGAWeedonMNBewsheaCMHLA-DQA1-HLA-DRB1 variants confer susceptibility to pancreatitis induced by thiopurine immunosuppressantsNat Genet2014461131113425217962
- LennardLTPMT in the treatment of Crohn’s disease with azathioprineGut20025114314612117866
- DubinskyMCLamotheSYangHYPharmacogenomics and metabolite measurement for 6-mercaptopurine therapy in inflammatory bowel diseaseGastroenterology200011870571310734022
- WeinshilboumRMSladekSLMercaptopurine pharmacogenetics: monogenic inheritance of erythrocyte thiopurine methyltransferase activityAm J Hum Genet1980326516627191632
- MoonWLoftusEVJrReview article: recent advances in pharmacogenetics and pharmacokinetics for safe and effective thiopurine therapy in inflammatory bowel diseaseAliment Pharmacol Ther Epub2016214
- FongSCBlakerPAArenas-HernandezMMarinakiAMSandersonJDGetting the best out of thiopurine therapy: thiopurine S-methyltransferase and beyondBiomark Med20159516525605455
- StoccoGPelinMFrancaRPharmacogenetics of azathioprine in inflammatory bowel disease: a role for glutathione-S-transferase?World J Gastroenterol2014203534354124707136
- StoccoGMartelossiSBarabinoAGlutathione-S-transferase genotypes and the adverse effects of azathioprine in young patients with inflammatory bowel diseaseInflamm Bowel Dis200713576417206640
- DervieuxTBoulieuRSimultaneous determination of 6-thioguanine and methyl 6-mercaptopurine nucleotides of azathioprine in red blood cells by HPLCClin Chem1998445515559510860
- StoccoGCuzzoniEDe IudicibusSDeletion of glutathione-S-transferase M1 reduces azathioprine metabolite concentrations in young patients with inflammatory bowel diseaseJ Clin Gastroenterol201448435123787247
- DubinskyMCLamotheSYangHYPharmacogenomics and metabolite measurement for 6-mercaptopurine therapy in inflammatory bowel diseaseGastroenterology200011870571310734022
- OstermanMTKunduRLichtensteinGRLewisJDAssociation of 6-thioguanine nucleotide levels and inflammatory bowel disease activity: a meta-analysisGastroenterology20061301047105316618398
- HindorfULindqvistMHildebrandHFagerbergUAlmerSAdverse events leading to modification of therapy in a large cohort of patients with inflammatory bowel diseaseAliment Pharmacol Ther20062433134216842460
- YarurAJAbreuMTDeshpandeARKermanDHSussmanDATherapeutic drug monitoring in patients with inflammatory bowel diseaseWorld J Gastroenterol2014203475348424707130
- Prieto-PérezRAlmogueraBCabaleiroTHakonarsonHAbad-SantosFAssociation between genetic polymorphisms and response to anti-TNFs in patients with inflammatory bowel diseaseInt J Mol Sci20161722526861312
- HlavatyTPierikMHenckaertsLPolymorphisms in apoptosis genes predict response to infliximab therapy in luminal and fistulizing Crohn’s diseaseAliment Pharmacol Ther20052261362616181301
- MoroiREndoKKinouchiYFCGR3A-158 polymorphism influences the biological response to infliximab in Crohn’s disease through affecting the ADCC activityImmunogenetics20136526527123358932
- KoderSRepnikKFerkoljIGenetic polymorphism in ATG16l1 gene influences the response to adalimumab in Crohn’s disease patientsPharmacogenomics20151619120425712183
- JürgensMLaubenderRPHartlFDisease activity, ANCA, and IL23R genotype status determine early response to infliximab in patients with ulcerative colitisAm J Gastroenterol20101051811181920197757
- BankSAndersenPSBurischJAssociations between functional polymorphisms in the NFB signaling pathway and response to anti-TNF treatment in Danish patients with inflammatory bowel diseasePharmacogenom J201414526534
- FujinoSAndohABambaSIncreased expression of interleukin 17 in inflammatory bowel diseaseGut2013526570
- AfifWLoftusEVJrFaubionWAClinical utility of measuring infliximab and human anti-chimeric antibody concentrations in patients with inflammatory bowel diseaseAm J Gastroenterol20101051133113920145610
- BortlikMDuricovaDMalickovaKInfliximab trough levels may predict sustained response to infliximab in patients with Crohn’s diseaseJ Crohns Colitis2013773674323200919
- Van MoerkerckeWAckaertCCompernolleGHigh infliximab trough levels are associated with mucosal healing in Crohn’s diseaseGastroenterology2010138S60
- BaertFNomanMVermeireSInfluence of immunogenicity on the long-term efficacy of infliximab in Crohn’s diseaseN Engl J Med200334860160812584368
- RoblinXMarotteHRinaudoMAssociation between pharmacokinetics of adalimumab and mucosal healing in patients with inflammatory bowel diseasesClin Gastroenterol Hepatol201412808423891927
- YarurAJJainAHauensteinSIHigher adalimumab levels are associated with histologic and endoscopic remission in patients with Crohn’s disease and ulcerative colitisInflamm Bowel Dis20162240941526752470
- SeowCHNewmanAIrwinSPSteinhartAHSilverbergMSGreenbergGRTrough serum infliximab: a predictive factor of clinical outcome for infliximab treatment in acute ulcerative colitisGut201059495419651627
- KarmirisKPaintaudGNomanMInfluence of trough serum levels and immunogenicity on long-term outcome of adalimumab therapy in Crohn’s diseaseGastroenterology20091371628164019664627
- NandaKSCheifetzASMossACImpact of antibodies to infliximab on clinical outcomes and serum infliximab levels in patients with inflammatory bowel disease (IBD): a meta-analysisAm J Gastroenterol2013108404723147525
- VelayosFSSheibaniSLocktonSPrevalence of antibodies to adalimumab (ATA) and correlation between ATA and low serum drug concentration on CRP and clinical symptoms in a prospective sample of IBD patientsGastroenterology2013144S91
- TewGWHackneyJAGibbonsDAssociation between response to etrolizumab and expression of integrin αE and granzyme A in colon biopsies of patients with ulcerative colitisGastroenterology201615047748726522261
- SwidsinskiALadhoffAPernthalerAMucosal flora in inflammatory bowel diseaseGastroenterology2002122445411781279
- PhilpottDJGirardinSECrohn’s disease-associated Nod2 mutants reduce IL10 transcriptionNat Immunol20091045545719381138
- FrankDNRobertsonCEHammCMDisease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseasesInflamm Bowel Dis20111717918420839241
- RauschPRehmanAKunzelSColonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (secretor) genotypeProc Natl Acad Sci U S A2011108190301903522068912
- LuppCRobertsonMLWickhamMEHost-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of EnterobacteriaceaeCell Host Microbe2007211912918005726
- Darfeuille-MichaudABoudeauJBuloisPHigh prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s diseaseGastroenterology200412741242115300573
- SokolHLepagePSeksikPDoreJMarteauPTemperature gradient gel electrophoresis of fecal 16S rRNA reveals active Escherichia coli in the microbiota of patients with ulcerative colitisJ Clin Microbiol2006443172317716954244
- OhkusaTSatoNOgiharaTMoritaKOgawaMOkayasuIFusobacterium varium localized in the colonic mucosa of patients with ulcerative colitis stimulates species-specific antibodyJ Gastroenterol Hepatol20021784985312164960
- OhkusaTYoshidaTSatoNWatanabeSTajiriHOkayasuICommensal bacteria can enter colonic epithelial cells and induce proinflammatory cytokine secretion: a possible pathogenic mechanism of ulcerative colitisJ Med Microbiol20095853554519369513
- AtarashiKTanoueTOshimaKTreg induction by a rationally selected mixture of Clostridia strains from the human microbiotaNature201350023223623842501
- LlopisMAntolinMCarolMLactobacillus casei downregulates commensals’ inflammatory signals in Crohn’s disease mucosaInflamm Bowel Dis20091527528318839424
- SokolHPigneurBWatterlotLFaecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patientsProc Natl Acad Sci U S A2008105167311673618936492
- WillingBHalfvarsonJDicksvedJTwin studies reveal specific imbalances in the mucosa associated microbiota of patients with ileal Crohn’s diseaseInflamm Bowel Dis20091565366019023901
- VarelaEManichanhCGallartMColonisation by Faecalibacterium prausnitzii and maintenance of clinical remission in patients with ulcerative colitisAliment Pharmacol Ther20133815116123725320
- PascalVPozueloMBorruelNA microbial signature for Crohn’s diseaseGut201766581382228179361
- MoayyediPSuretteMGKimPTFecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trialGastroenterology20151102109
- RossenNGFuentesSvan der SpekMJFindings from a randomized controlled trial of fecal transplantation for patients with ulcerative colitisGastroenterology20151110118