Abstract
RNA profiling is increasingly used to predict drug response, dose, or toxicity based on analysis of drug pharmacokinetic or pharmacodynamic pathways. Before implementing multiplexed RNA arrays in clinical practice, validation studies are carried out to demonstrate sufficient evidence of analytic and clinical performance, and to establish an assay protocol with quality assurance measures. Pathologists assure quality by selecting input tissue and by interpreting results in the context of the input tissue as well as the technologies that were used and the clinical setting in which the test was ordered. A strength of RNA profiling is the array-based measurement of tens to thousands of RNAs at once, including redundant tests for critical analytes or pathways to promote confidence in test results. Instrument and reagent manufacturers are crucial for supplying reliable components of the test system. Strategies for quality assurance include careful attention to RNA preservation and quality checks at pertinent steps in the assay protocol, beginning with specimen collection and proceeding through the various phases of transport, processing, storage, analysis, interpretation, and reporting. Specimen quality is checked by probing housekeeping transcripts, while spiked and exogenous controls serve as a check on analytic performance of the test system. Software is required to manipulate abundant array data and present it for interpretation by a laboratory physician who reports results in a manner facilitating therapeutic decision-making. Maintenance of the assay requires periodic documentation of personnel competency and laboratory proficiency. These strategies are shepherding genomic arrays into clinical settings to provide added value to patients and to the larger health care system.
Introduction
RNA profiling supplements traditional histopathologic, immunologic, cytogenetic, and proteomic means of pharmacogenetic analysis.Citation1 By testing tens to thousands of RNAs at once, signatures are generated that reflect abundant and also redundant data on clinical status that could provide added value beyond what is achieved by testing a single analyte. In addition to testing messenger RNA, emerging data on noncoding RNA expression (microRNAs and long noncoding RNAs) represents a new frontier for expression profiling that is likely to inform patient management decisions further.Citation2
RNA panels are increasingly being adopted in clinical trials and ultimately, once vetted as reliable and useful, in routine health care settings for decision-making about drug efficacy, to monitor drug action in the intended biochemical pathway or in off-target pathways, or to select optimal dosage. Reliable RNA profiling builds on the same quality assurance principles that have guided laboratory medicine over the past few decades. Among the many factors contributing to good outcomes are personnel competency with demonstrated proficiency in achieving expected results, and quality control methods to detect deficiencies in an assay or in the specimen being assayed. Quality of RNA-based profiling has improved over the past decade as a result of several factors, ie, good manufacturing practices making available standardized reagents, controls, and instrumentation,Citation3,Citation4 biospecimen research demonstrating best practices to process tissue and to preserve RNA,Citation5,Citation6 novel paradigms for quality control to assess analytic performance of the signature rather than of individual components, and software presenting control and patient data to laboratory scientists in a manner facilitating analytic and clinical interpretation.Citation7
Assay validation
Quality assurance parameters are established and refined during validation studies. The main goal of validation work is to demonstrate whether an assay is analytically sound, clinically useful, and of sufficient added value to deem it medically necessary for the care of a defined group of patients. Assay validation guidance published by the College of American Pathologists suggest that studies should be carried out in three parts, ie, a planning phase to devise the assay for its intended use, a data collection phase to gather results on analytic and clinical performance characteristics (eg, sensitivity, specificity), and an implementation phase to transition the assay to the clinical setting once the assay is vetted by the clinical laboratory director. Key steps are summarized in .Citation8 The validation study defines acceptable specimen types, indications for testing, and a standard operating procedure for performing the test,Citation8–Citation10 to include pertinent quality checks and controls.Citation11,Citation12
Translational research teams seeking to validate array-based assays should include technology specialists, clinical trial experts, statisticians, clinicians who will order the test and act on test results, and clinical laboratory technicians who perform the assays and interpret the results. A multi-disciplinary development team increases the likelihood of producing an assay that is practical, robust, and sufficiently useful to be incorporated into routine patient care.Citation13
A rule of thumb for successful assay design is to use the simplest and safest strategy that meets the clinical objective. For example, Microarray Quality Control (MAQC) project data suggests that a one-color approach yields results equivalent to a two-color approach, while also using fewer reagents and eliminating the potential for bleed-through of fluorochrome from one channel to the other.Citation14–Citation16 Thoughtful assay design can help minimize problematic delays that ensue when a critical instrument undergoes repair, when a reagent is on “back-order”, or when a patient test must be repeated. This article describes strategies to assure quality of RNA profiling during assay validation and ultimately in day-to-day laboratory medicine practice. This quality assurance work is meant to ward off error and to detect and correct problems when they occur.
Specimen collection, handling, and storage
Most errors in clinical laboratory assays occur in the preanalytic phase of testing, emphasizing the importance of validating the assay on real-world conditions for specimen collection and transport, processing, and storage prior to analysis.Citation17–Citation21 According to some reports, the variables confounding RNA signatures can be quite unexpected, such as whether the first or second needle biopsy was tested,Citation22 or whether cells were frozen prior to analysis.Citation23,Citation24 It is important that validation work address preanalytic variables as a component of the overall efficacy of an assay.Citation6,Citation25–Citation27
RNA tends to be unstable, and some transcripts rapidly degrade under adverse collection, storage, or handling conditions.Citation25 Stabilization of RNA at the time of whole blood collection is achievable using commercial collection vials,Citation28–Citation33 although the benefits of bedside stabilization must be weighed against the downsides that include stocking special blood collection tubes in every applicable blood collection station, and inability to use cell separation technologies to separate subpopulations of white cells or to eliminate unwanted erythrocytes containing abundant globin RNA.
Criteria for acceptance or rejection of specimens should be established during assay validation.Citation34–Citation36 In solid tissue, the pathologist who selects tissue for analysis follows a protocol specifying acceptability criteria, such as the minimum proportion of cells that must be malignant in order to generate the relevant tumor-related signatures.Citation37 Interpretation of downstream molecular results is done in the context of the input tissue.
Prolonged fixation in formalin causes RNA crosslinking and thwarts RNA extraction.Citation38,Citation39 However, formalin fixation also prevents tissue degradation and reduces or eliminates RNase function, permitting such tissues to be profiled successfully after histopathologic examination and storage.Citation40–Citation42 Chung et al showed that fixation times of 4–48 hours were reasonable, with 12–24 hours yielding the best RNA in downstream analysis.Citation43 For some target RNAs, expression is similar in paraffin blocks and matched frozen tissue,Citation44–Citation47 but other RNA targets do not correlate as well.Citation47,Citation48
A molecular test is most likely to be adopted if its input specimen type is formalin-fixed, paraffin-embedded tissue, recognizing that variation in the age of a tissue block and its processing variables must also be considered as factors impacting RNA profile.Citation34 RNA quality is better in frozen tissue than in formalin fixed tissue; however frozen tissue is not usually saved in clinical settings because of limited tissue volume, upfront uncertainty about the extent of sampling or testing needed to make a traditional histopathologic diagnosis, and the cost of saving and storing tissue by nonstandard methods when most such tissue will not meet the indications for any downstream test.
Acid decalcification, a process typically done to permit histologic sectioning of bony tissues, causes depurination with fragmentation of nucleic acid. Therefore, bone marrow aspirate or clot sections may be more suitable for molecular testing than is a marrow biopsy. Alcohol-based fixatives may yield higher quality RNA than is achieved by formalin fixation,Citation49–Citation51 but alternative fixatives are inferior with respect to histologic detail required for a pathologist’s microscopic diagnosis and also adverse impact on immunostains that are frequently applied in diagnostic workup of solid tissues.Citation52 An important preanalytic step involves assurance that the input tissue is representative of the organ or lesion being evaluated.Citation37 Pathologist vetting of tissue acceptability is done by microscopic examination of a stained slide (eg, frozen section, paraffin section, cytologic preparation, or smear). When cell enrichment is required in blood or marrow, flow cytometry or magnetic bead separation is applied.Citation53 In solid tissue, cell enrichment is done by macrodissection or microdissection.Citation54–Citation59
Controls, quality checks, and limits on their acceptability
Compared with individual tests, array-based RNA profiles create novel challenges for quality control and for data interpretation.Citation60 While traditional single-analyte assays require inclusion of a positive and a negative control in every run, it is clear that microarray tests cannot possibly include a separate control for each of the tens to thousands of target analytes. Thus, a new paradigm of quality control has emerged to accommodate array-based testing by demonstrating that the resulting RNA signature is accurate and reproducible.
Each control is run alongside the patient specimens to generate a result that must fall within previously established limits. Aliquots of residual natural human tissue may be heterogeneous but they are still among the best controls because they closely resemble patient specimens and they can be included in all steps of the assay. However, it is difficult to obtain large amounts of residual patient specimens, so xenograft tissue is a suitable alternative. Stored residual RNA from previously tested blood or tissue specimens is also a suitable control. Cell lines are useful because they can be diluted to test sensitivity and linearity of the test system, and typically these cells or their derivatives are spiked into appropriate matrix so as to mimic patient specimens as closely as possible. A mixture of ten cell lines is used to prepare the Agilent/Stratagene reference RNA that is well characterized in multiple sample exchange studies.Citation57,Citation61,Citation62
Controls are designed to test critical aspects of assay performance and to help pinpoint sources of error. Several types of controls are used, ie, “no template” controls evaluate background noise and detect contamination by stray nucleic acid. Exogenous controls are run alongside patient specimens in all or in selected stages of the stepwise protocol to check generic assay performance. In an assay with five major outcome groups, it is reasonable to rotate exogenous controls so that any given run contains a control for at least one of the outcome groups (see ). A failed control would then trigger investigation of all runs since that control last passed muster.
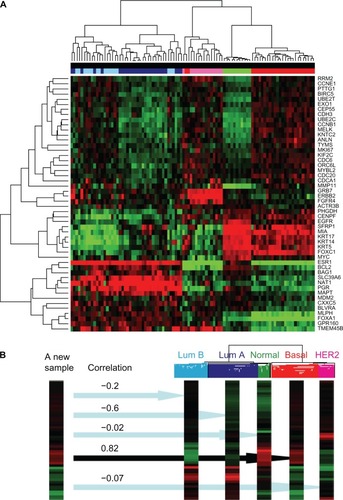
Figure 2 Data interpretation is facilitated by software manipulation of abundant data generated by profiling virtually all approximately 22,000 human genes. (A) An unsupervised clustering algorithm was applied to the full dataset and then to a subset of 50 RNAs listed on the right to generate a heatmap showing patterns of expression in 96 breast cancer tissues. (B) A single sample predictor algorithm helps assign a subtype to a new patient specimen using Spearman’s correlation coefficients to estimate certainty of the classification. The expression pattern of a new sample matches the basal subtype of breast cancer most closely, and this result is likely to influence clinical management by virtue of a poor prognosis and lack of response to traditional antineoplastic agents. Typically basal subtype tumors are termed “triple-negative” because they lack immunohistochemical expression of ESR1, PGR, and ERBB2 proteins, so parallel testing of such proteins might serve as a quality assurance measure for the analytic process and also for the clinical categorization of this patient’s disease.
Endogenous controls measure elements that are inherent to a given patient specimen, such as a housekeeping transcript level to address cell viability, cellularity, transport, processing, storage, and RNA extraction steps. An RNA signature typically tests for multiple housekeeping transcripts that were selected during validation work for their consistent amount (low, moderate, or high) in the pertinent specimen type. In routine testing, their expression is an indicator of hybridizable RNA that permits rejection of specimens with inadequate RNA quality.Citation45,Citation63 Expression levels of one or more housekeepers could serve as a normalizer by which to gauge expression of other transcripts.Citation64–Citation66
Spiked controls can evaluate assay performance, at least for those steps of analysis after spiking occurs. Commercial RNA spikes of known sequence (developed by the External RNA Controls Consortium) can be added to each patient specimen either at the time that lysis buffer is added or later when RNA is being prepared for analysis.Citation67–Citation71 Their downstream measurement can detect interfering substances such as autofluorescence, heparin anticoagulant, hemoglobin protein or globin RNA, or residual phenol. To track specimens through the many steps of specimen preparation and analysis, combinations of spiked molecules have been proposed as specimen identifiers.Citation72
Any control result or quality check falling outside acceptable limits is investigated for the cause of the failure, so that corrective action may be taken when feasible. For example, if spectrophotometry indicates failure of all specimens in a given run, including the control, the extraction procedure is likely to be the culprit. On the other hand, adequacy of the control and all but one of the patients in a given run would indicate which patient specimen to reject or re-extract. Failed hybridization of spiked controls or housekeeping transcripts could help pinpoint whether the flaw lies before or after spiking, thus impacting the action plan in response to the aberrant result. Control results are always documented, as are the actions taken in response to a failure, to promote quality improvement over time.
RNA quality and hybridization reactions
Automated instruments promote standardization of blood or tissue RNA extractions and also reduce labor costs. The choice of extraction method can impact an RNA signature,Citation73 confirming the need to validate the extraction method in concert with the rest of the test system. RNA quantity is often measured using ultraviolet spectrophotometry or fluorimetry, keeping in mind that any DNA interferes with RNA measurement. RNA size may be visualized by electrophoresis, and software algorithms such as the RNA integrity number score have been developed to grade RNA quality.Citation63,Citation66 Although delayed processing by up to an hour does not adversely affect the RNA integrity number score, it can affect the RNA signature.Citation35,Citation52,Citation74–Citation77
Linear preamplification of RNA permits analysis of very small specimens and also can incorporate a label to permit RNA detection in downstream analysis.Citation45,Citation55,Citation78–Citation81 In preparation for the reverse transcription polymerase chain reaction, genomic DNA is usually removed from nucleic acid extracts prior to cDNA preparation. Since aRNA or cDNA preparation can introduce bias, some scientists suggest performing replicates, however the work required to resolve discrepant findings implies that replicate testing will not overcome deficiencies in a poorly designed assay. Clinical grade assays must be robust enough that significant variance in RNA signatures between two patients largely represents biological difference rather than technical error.
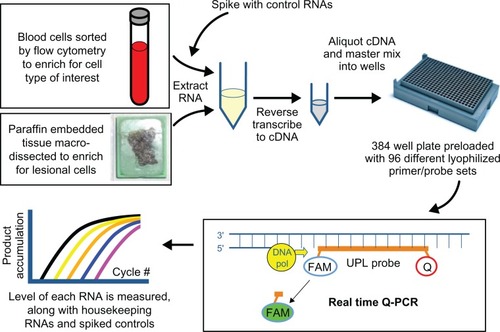
Quantitative reverse transcription polymerase chain reaction has a long track record in clinical laboratories, and high throughput quantitative reverse transcription polymerase chain reaction systems are capable of measuring tens to hundreds of RNAs at onceCitation82 (see ). Denser array platforms, such as Affymetrix and Agilent microarray systems, are also gaining ground as quality concerns are successfully addressed.Citation2,Citation14,Citation83–Citation86 On the horizon are full transcriptome sequencing technologies.Citation85,Citation87
Figure 3 RNA profiling by high throughput quantitative reverse transcription polymerase chain reaction is a complex process requiring multiple quality checks. In this example, cell enrichment procedures are applied to blood or tissue on which a pathologist has confirmed lesional cells, spiked control RNA is added during cell lysis to measure downstream assay performance in the patient specimen, spectrophotometry assures adequate recovery of purified RNA, robotics standardize loading of cDNA and master mix into 384-well Roche RealTime Ready plates preloaded at the factory with lyophilized primers and probes, an internal probe enhances specificity of amplicon measurement beyond that achieved with dye alone, housekeeping RNA measurements reflect the overall adequacy of the specimen and the test system, and decision-support software assists with laboratory physician interpretation of the findings.
Manufacturers are crucial for providing reagents, instruments, and chips to testing laboratories. Clinical laboratories tend to choose manufacturers complying with Food and Drug Administration good manufacturing practices or equivalent International Standardization Organization programs promoting quality and consistency of product across lot numbers.Citation3,Citation4,Citation88,Citation89 Manufacturers’ products are additionally vetted by the testing laboratory to assure adequate performance for their intended use.
Hybridization reactions are subject to error because of cross-reactivity, interference due to secondary structure or dimerization diminishing intended base pairing, and competition between two or more simultaneous reactions in a single vessel. Many of these concerns are addressed during the validation study conducted prior to clinical implementation. An advantage of array-based assays is the potential to provide redundancy by targeting the same analyte numerous times, such as testing it in different physical quadrants of an array, or targeting several conserved segments of the same transcript using 3′, 5′, and intermediary probes. In the virology realm, one could target multiple conserved segments of an RNA viral genome. If a certain biochemical pathway or phenotype is critical, one could target multiple markers signifying that pathway or phenotype. In these scenarios, one capitalizes on the strength of the array in simultaneous analyses.
Data analysis and interpretation
Data interpretation is done in the context of a thorough understanding of the technical strengths and weaknesses of the test system, as well as medical issues relevant to the dilemma that the test is meant to solve, building on expertise and prior experience gathered during the validation study and in subsequent clinical practice. Because raw data from massive parallel testing can be quite abundant, software algorithms must present selected data in a manner that facilitates interpretation.Citation7 A protocol is followed to generate the dataset for interpretation, such as applying a normalization strategy to adjust for background, or log transformation to facilitate comparison with other samples.Citation64,Citation65,Citation90–Citation93 To avoid masking the very biologic variability that is being evaluated, excessive manipulation of data should be avoided.Citation94
There are two phases of interpretation, ie, analytic and clinical. Analytic interpretation involves generating a reportable result after first evaluating selected data on the controls and on the patient. Clinical interpretation conveys the significance of the result in patient management. After applying pertinent software algorithms, a package of data, both raw and processed, is assembled for review by the interpreting pathologist or laboratory scientist.
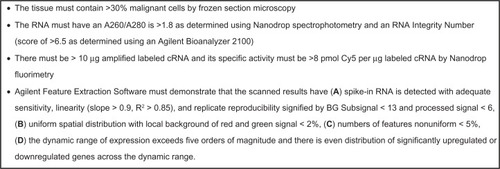
The first step in analytic interpretation is to review results of controls and quality checks. For frozen tissue profiling, example quality checks are listed in . For paraffin embedded tissue profiling, Roberts et al used reverse transcription polymerase chain reaction with an ACTB 3′ to 5′ ratio of <20, a Ct of <7 between ACTB 5′ and the Agilent/Stratagene Universal Reference RNA, and a 28 s rRNA delta Ct <15 to vet RNA before Affymetrix profiling.Citation48
Figure 4 Example quality checks on frozen tissue profiled using an Agilent microarray two-color strategy. This is an example; acceptance limits must be established for each application.
Controls of critical importance are developed during validation studies and applied in routine clinical testing. For example, melanoma tissue frequently expresses several melanocyte-specific genes, such as MAGEA1, MITF, MART1 (melan-A), CMM [HMB45], S100, and TYR (tyrosinase), and expression of these factors is a reasonable reflection of the presence of melanocytic cells.
When redundant assays are present on the array, replicates are examined for consistency or to find problematic variations. When redundant pathways or functions are evaluated using separate probes, trends tend to promote confidence in an interpretation, while inconsistent results are a red flag for a technical problem versus patient-specific variation. For example, a highly proliferative tumor is expected to overexpress most of the known proliferation markers on the array.
Criteria for vetting data generated using Affymetrix arrays have been proposed by Staal et al.Citation73 These include examining the 3′ to 5′ ratio for selected housekeeping genes (1 is ideal, <3 is good), assuring the dynamic range of the output signals, checking uniformity across the array chip, and determining if number of expressed RNAs exceeds 25% of the total.
To categorize the findings in a given patient, a predictive model may be applied that finds patterns across many analytes, as facilitated by a “single sample predictor” algorithm.Citation46,Citation83,Citation95–Citation99 The ability to view global patterns of gene expression is a unique strength of expression profiling compared with the discrete testing that characterizes traditional laboratory analysis. An assignment is typically accompanied by a statistic (eg, Spearman’s correlation coefficients) representing the strength of the match to one diagnostic category versus each of the others.Citation100
A clustering program and a heat map may be generated to display graphically the results of a given patient in a dendrogram alongside the patients in the training and validation sets, to help categorize the patient into pre-established groups based on similarity of expression pattern (see ). Caution is required when a profile falls at the border between two groups since that patient may not belong to either group. Computer generated scores or predictors should be checked to assure they make sense based on evaluation of pertinent raw data. Assessing the degree of confidence in the result is helpful for downstream clinical interpretation and reporting described below.
Clinical interpretation of results, reporting, and consultations
Clinical interpretation is done in the context of the clinical indication for which the test was ordered. In this regard, the laboratory test order represents a request for consultation in which the laboratory physician’s interpretation answers the question posed by the physician who ordered the test. Even if the end result is a numeric score or a discrete disease classification, it is helpful to interpret the result in light of the input tissue characteristics, pertinent limitations of the assay based on quality checks that were performed during analysis, and the level of confidence in the result. Additional correlative analysis may be done using patient information that is independent of the data generated during testing (eg, age, gender, tumor stage, immunohistochemical, or flow cytometric findings). Most importantly, the impact on clinical decision-making should be described, along with any recommended follow-up. For example, a lymphoblastic leukemia patient whose profile matches the BCR-ABL1 group implies pharmacogenetic response to tyrosine kinase inhibitor therapy, such as desatinib or nilotinib.Citation101,Citation102 This classification also suggests the need to confirm that the translocation (p210 versus p190 breakpoint) is amplifiable by quantitative reverse transcription polymerase chain reaction for purposes of monitoring disease burden during therapy.Citation102
Finally, it should be noted that pathologists and other laboratory scientists are accustomed to dealing with unexpected findings. After all, interpreting histologic slides or karyotypes are examples of open-ended procedures for which results may turn out to be completely different from the suspected diagnosis for which the test was ordered. Examination of expression data may yield alternative interpretations that complement or override the objective data generated by a software algorithm. Medical judgment is needed to decide which data are reportable, and to describe the clinical significance of relevant findings. Decisions should be based on technical and medical evidence from published literature, validation work, and other reliable sources such as databases of expression profiles on patients of known diagnosis or outcome.Citation103,Citation104
The report placed in the patient’s medical record contains a written summary of the results and an interpretation that facilitates subsequent decision-making, as recommended in the College of American Pathologists’ guidance for molecular test reporting.Citation105 Composing a report that is concise yet informative requires technical and medical training, as well as attention to detail. Quality assurance measures might include review of the report for transcription error, review of the raw data and interpretation by a different medical professional, and review of medical records to assure transmission with appropriate formatting.Citation106
Data storage and retrospective mining
Custom-designed and off-the-shelf arrays are available from multiple manufacturers. If an off-the-shelf chip is used, then software can be programmed to mask irrelevant data. United States regulations call for results to be stored for five years in a manner protecting privacy and data integrity. Archival versions of the procedure manual serve to annotate each dataset by linking to the methods used to create the data. It is feasible that the array dataset could be used for one indication at the time of initial testing, and for other indications later (eg, first a diagnostic test, then a prognostic test, then several predictive tests during the course of first-line and second-line therapy selections). The process of revisiting the same patient dataset over and over is analogous to reviewing microscopic slides again in the context of new histopathologic criteria for diagnosis or newly available histochemical assays.
Government regulation and guidance from professional groups
When laboratory test results are used to guide patient management, even in the context of a clinical trial, then the results must be reliable. In the United States, all such tests are performed in laboratories meeting regulatory standards codified in the Clinical Laboratory Improvement Amendments. Manufacturers of reagents and devices are subject to regulations governing the Food and Drug Administration. Many pharmacogenetic tests have been approved by the Food and Drug Administration, including those targeting RNA of microbial organisms (eg, hepatitis C virus, human immunodeficiency virus, mycobacteria, influenza and other respiratory viruses) and tests for cancer (Agendia Mammaprint,Citation107–Citation109 Pathwork Diagnostics Tissue of Origin Test,Citation100 and a Veridex assay that is no longer marketed) and transplant rejection (xDx AlloMap). Examples of RNA-based pharmacogenetic tests that were developed and validated in individual testing laboratories include the BCR-ABL1 transcript levels and ABL1 mutation status to predict efficacy or dose of tyrosine kinase inhibitor therapy,Citation110 and Genomic Health’s Oncotype Dx assay for which a recurrence score influences decision-making about use of chemotherapy in breast cancer patients.Citation111,Citation112 Pathologists and other physicians in each high complexity testing laboratory are responsible for assuring that tests meet regulatory standards and that appropriate medical consultation is available to clients.Citation113 To meet regulatory guidelines in the United States, it is recommended that a physician with molecular subspecialty board certification, document the suitability of the quality control work by signing both the procedure manual and the assay validation report associated with any laboratory developed test.
The MAQC is a Food and Drug Administration initiative addressing the quality of RNA-based microarray expression profiling.Citation114 Interlaboratory exchanges of samples and datasets showed that RNA analysis is technically robust as are the bioinformatic prediction models for categorizing array datasets.Citation61,Citation115–Citation118 Several clinical professional groups have developed standards for RNA-based testing services, including the laboratory accreditation program of the College of American Pathologists that provides checklists serving as a roadmap for high quality molecular testing,Citation119 and the Clinical and Laboratory Standards Institute that has dozens of documents describing standards for validating, implementing, and maintaining molecular assays. Examples include diagnostic nucleic acid microarrays,Citation96 use of external RNA controls in gene expression assays,Citation120 and verification and validation of multiplex nucleic acid assays.Citation121 Helpful guidance is also found in a European guideline for RNA signatures in leukemiaCitation73 and in clinical pharmacogenetic testing guidelines from the National Academy of Clinical Biochemistry.Citation122
Personnel competency and laboratory proficiency
Perhaps the single most important factor in assuring a good outcome is the personnel competency, beginning with the clinician who orders the test and proceeding to those who collect, transport, and handle specimens, followed by those who perform, interpret, and act on test results. Meticulous care is required to avoid RNA degradation by using RNAse-free materials, frequently changing gloves and bench covers, and using 10% bleach or RNaseZap to eliminate extraneous nucleic acid from surfaces. Standard clinical-grade work processes include assuring functionality of each new lot number of reagent prior to its use in patient care, routine preventive maintenance with function checks for each instrument, and competency checks of technical personnel after training and before initiating patient testing, and again on a periodic basis.
Generating an RNA signature requires multistep transfers of a specimen or its derivative which in turn requires painstaking effort to maintain specimen integrity and identification.Citation20,Citation123,Citation124 Robotic systems can potentially standardize pipetting and transfer, and barcodes facilitate specimen tracking and labeling.Citation124,Citation125 Robots should be programmed to minimize the risk of carryover and contamination.
Proficiency surveys challenge the testing laboratory’s performance, educate laboratory personnel, and encourage improvement.Citation126–Citation129 Such surveys involve periodic analysis of “unknown” specimens followed by an evaluation of performance against other laboratories doing similar assays. Formal proficiency surveys are offered for some RNA-based pharmacogenetic tests, such as HIV genotyping and PML-RARA translocation. While no formal survey exists for expression profiling, proof of concept that array-based testing is amenable to proficiency testing is shown by the College of American Pathologists’ cytogenomic microarray survey which supports interlaboratory comparisons for DNA-based gene copy number analysis. Alternative assessment methods include exchanging samples with laboratory that performs similar tests, or retesting internal samples as if they were unknowns.Citation130 The Association for Molecular PathologyCitation131 and the GeneTests websiteCitation132 maintain directories of testing laboratories, and the College of American Pathologists can also help identify a laboratory with whom to exchange specimens.Citation119
Conclusion
RNA profiling is increasingly used to substantiate drug selection or dosage. In the infectious disease realm, molecular analysis of microbial genomes and drug resistance factors can accelerate the time to results and powerfully predict antimicrobial drug efficacy. In the oncology arena, RNA signatures may provide added value for selecting a drug regimen that is likely to overcome the biochemical defect(s) driving tumor cell proliferation.Citation133,Citation134 Serial testing is being explored as a way to document the impact of the drug regimen in the intended biochemical pathway or in off-target pathways.Citation135
The strategies for quality assurance described herein have shepherded expression profiling into clinical settings. With special attention to RNA quality and data analysis tools, it is likely that robust, accurate, and reproducible RNA-based assays will continue to be developed and implemented. These assays are powerful by virtue of the number of RNAs and pathways that are evaluated, and by redundancy that boosts confidence in the findings.
Acknowledgements
The authors thank Charles Perou, Greg Ray, Karen Weck, Jessica Booker, Myla Lai-Goldman, and Yan Li of the University of North Carolina at Chapel Hill. This work was supported by the Department of Pathology and Laboratory Medicine, the University Cancer Research Fund, and two grants to the University of North Carolina at Chapel Hill from the National Institutes of Health including a Clinical Translational Science Award and funds from the Alliance for Clinical Trials in Oncology.
Disclosure
MLG is a consultant for McKesson, Abbott Laboratories, Roche Molecular Diagnostics, and serves on the clinical advisory board of Generation Health.
References
- Tumor Analysis Best Practices Working GroupExpression profiling – best practices for data generation and interpretation in clinical trialsNat Rev Genet20045322923714970825
- AuerHNewsomDLKornackerKExpression profiling using Affymetrix GeneChip microarraysMethods Mol Biol2009509354619212713
- FuscoeJCTongWShiLQA/QC issues to aid regulatory acceptance of microarray gene expression dataEnviron Mol Mutagen200748534935317567852
- HackettJLGutmanSIIntroduction to the Food and Drug Administration (FDA) regulatory processJ Proteome Res2005441110111316083260
- ImbeaudSAuffrayCThe ‘39 steps’ in gene expression profiling: Critical issues and proposed best practices for microarray experimentsDrug Discov Today200510171175118216182210
- National Cancer InstituteNCI Best Practices for Biospecimen ResourcesBethesda, MDUnited States National Cancer Institute2010 Available from: http://www.allirelandnci.com/pdf/NCI_Best_Practices_060507.pdf. Accessed August 18, 2011.
- FoxPHendlerJChanging the equation on scientific data visualizationScience2011331601870570821311008
- JenningsLVan DeerlinVMGulleyMLRecommended principles and practices for validating clinical molecular pathology testsArch Pathol Lab Med2009133574375519415949
- BurdEMValidation of laboratory-developed molecular assays for infectious diseasesClin Microbiol Rev201023355057620610823
- KohlMDevelopment and validation of predictive molecular signaturesCurr Mol Med201010217317920196729
- ArcherKJMasVRO’BrienTRPfeifferRLumNLFisherRAQuality assessment of microarray data in a multicenter studyDiagn Mol Pathol2009181344319214110
- BammlerTBeyerRPBhattacharyaSStandardizing global gene expression analysis between laboratories and across platformsNat Methods20052535135615846362
- CoyleVMJohnstonPGGenomic markers for decision making: What is preventing us from using markers?Nat Rev Clin Oncol201072909720010899
- ShackSGene expression profiling of tissues and cell lines: A dual-color microarray methodMethods Mol Biol201170012514321204031
- OberthuerAJuraevaDLiLComparison of performance of one-color and two-color gene-expression analyses in predicting clinical endpoints of neuroblastoma patientsPharmacogenomics J201010425826620676065
- PattersonTALobenhoferEKFulmer-SmentekSBPerformance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) projectNat Biotechnol20062491140115016964228
- HollandNTSmithMTEskenaziBBastakiMBiological sample collection and processing for molecular epidemiological studiesMutat Res2003543321723412787814
- De CeccoLMusellaVVeneroniSImpact of biospecimens handling on biomarker research in breast cancerBMC Cancer2009940919930681
- LippiGChanceJJChurchSPreanalytical quality improvement: From dream to realityClin Chem Lab Med20114971113112621517699
- PlebaniMErrors in clinical laboratories or errors in laboratory medicine?Clin Chem Lab Med200644675075916729864
- SchmittMMengeleKSchuerenEEuropean Organisation for Research and Treatment of Cancer (EORTC) Pathobiology Group standard operating procedure for the preparation of human tumour tissue extracts suited for the quantitative analysis of tissue-associated biomarkersEur J Cancer200743583584417321128
- DrubinDSmithJSLiuWZhaoWChaseGAClawsonGAComparison of cryopreservation and standard needle biopsy for gene expression profiling of human breast cancer specimensBreast Cancer Res Treat2005901939615770532
- JochumsenKMTanQDahlgaardJKruseTAMogensenORNA quality and gene expression analysis of ovarian tumor tissue undergoing repeated thaw-freezingExp Mol Pathol20078219510216842777
- VartanianKSlottkeRJohnstoneTGene expression profiling of whole blood: Comparison of target preparation methods for accurate and reproducible microarray analysisBMC Genomics2009101219123946
- RainenLArbiqueJCAsthanaDClinical Laboratory Standards Institute Document MM13-A: Collection, transport, preparation, and storage of specimens for molecular methods approved guidelineWayne, PAClinical and Laboratory Standards Institute2005
- Leyland-JonesBRAmbrosoneCBBartlettJRecommendations for collection and handling of specimens from group breast cancer clinical trialsJ Clin Oncol200826345638564418955459
- MooreHMKellyABJewellSDBiospecimen Reporting for Improved Study Quality (BRISQ)J Proteome Res20111083429343821574648
- WilliamsMAStabilizing the code-methods to preserve RNA prove their worthBiomark Insights2010513914321151590
- AsareALKolchinskySAGaoZDifferential gene expression profiles are dependent upon method of peripheral blood collection and RNA isolationBMC Genomics2008947418847473
- MathesonLADuongTTRosenbergAMYeungRSAssessment of sample collection and storage methods for multicenter immunologic research in childrenJ Immunol Methods20083391828918771669
- DebeySZanderTBrorsBPopovAEilsRSchultzeJLA highly standardized, robust, and cost-effective method for genome-wide transcriptome analysis of peripheral blood applicable to large-scale clinical trialsGenomics200687565366416387473
- ShouJDotsonCQianHROptimized blood cell profiling method for genomic biomarker discovery using high-density microarrayBiomarkers200510431032016191486
- WeberDGCasjensSRozynekPAssessment of mRNA and microRNA stabilization in peripheral human blood for multicenter studies and biobanksBiomark Insights201059510220981139
- HewittSMLewisFACaoYTissue handling and specimen preparation in surgical pathology: issues concerning the recovery of nucleic acids from formalin-fixed, paraffin-embedded tissueArch Pathol Lab Med2008132121929193519061293
- DashAMaineIPVaramballySShenRChinnaiyanAMRubinMAChanges in differential gene expression because of warm ischemia time of radical prostatectomy specimensAm J Pathol200216151743174812414521
- FerrueloAEl-AssarMLorenteJATranscriptional profiling and genotyping of degraded nucleic acids from autopsy tissue samples after prolonged formalin fixation timesInt J Clin Exp Pathol20114215616121326810
- RoepmanPde KoningEvan LeenenDDissection of a metastatic gene expression signature into distinct componentsGenome Biol2006712R11717156469
- Macabeo-OngMGinzingerDGDekkerNEffect of duration of fixation on quantitative reverse transcription polymerase chain reaction analysesMod Pathol200215997998712218216
- ChungJYHewittSMAn optimized RNA extraction method from archival formalin-fixed paraffin-embedded tissueMethods Mol Biol2010611192719960319
- BudcziesJWeichertWNoskeAGenome-wide gene expression profiling of formalin-fixed paraffin-embedded breast cancer core biopsies using microarraysJ Histochem Cytochem201159214615721339180
- SalehAZainRBHussainiHTranscriptional profiling of oral squamous cell carcinoma using formalin-fixed paraffin-embedded samplesOral Oncol201046537938620371203
- TonCCVartanianNChaiXGene expression array testing of FFPE archival breast tumor samples: An optimized protocol for WG-DASL sample preparationBreast Cancer Res Treat2011125387988320842525
- ChungJYBraunschweigTWilliamsRFactors in tissue handling and processing that impact RNA obtained from formalin-fixed, paraffin-embedded tissueJ Histochem Cytochem200856111033104218711211
- ParkerJSMullinsMCheangMCSupervised risk predictor of breast cancer based on intrinsic subtypesJ Clin Oncol20092781160116719204204
- AbduevaDWingMSchaubBTricheTDavicioniEQuantitative expression profiling in formalin-fixed paraffin-embedded samples by Affymetrix microarraysJ Mol Diagn201012440941720522636
- DuenwaldSZhouMWangYDevelopment of a microarray platform for FFPET profiling: Application to the classification of human tumorsJ Transl Med200976519638234
- MittempergherLde RondeJJNieuwlandMGene expression profiles from formalin fixed paraffin embedded breast cancer tissue are largely comparable to fresh frozen matched tissuePLoS One201162e1716321347257
- RobertsLBowersJSensingerKLisowskiAGettsRAndersonMGIdentification of methods for use of formalin-fixed, paraffin-embedded tissue samples in RNA expression profilingGenomics200994534134819660539
- CoxMLSchrayCLLusterCNAssessment of fixatives, fixation, and tissue processing on morphology and RNA integrityExp Mol Pathol200680218319116332367
- CoxMLEddySMStewartZSInvestigating fixative-induced changes in RNA quality and utility by microarray analysisExp Mol Pathol200884215617218291364
- LawsonMHRasslDMCummingsNMTissue banking of diagnostic lung cancer biopsies for extraction of high quality RNAJ Thorac Oncol20105795696320512072
- MedeirosFRiglCTAndersonGGBeckerSHHallingKCTissue handling for genome-wide expression analysis: A review of the issues, evidence, and opportunitiesArch Pathol Lab Med2007131121805181618081440
- HessCDenkersFOssenkoppeleGGene expression profiling of minimal residual disease in acute myeloid leukaemia by novel multiplex-PCR-based methodLeukemia200418121981198815470488
- WangSWangLZhuTImprovement of tissue preparation for laser capture microdissection: Application for cell type-specific miRNA expression profiling in colorectal tumorsBMC Genomics20101116320219115
- LuttgesJHahnSAHeidenblutAMManual microdissection combined with antisense RNA-longSAGE for the analysis of limited cell numbersMethods Mol Biol201057613515419882261
- MojicaWDSykesDEConroyJGaileDFangXNowakNA comparative analysis of two tissue procurement approaches for the genomic profiling of clinical colorectal cancer samplesInt J Colorectal Dis200823111089109818629512
- MojicaWDSteinLHawthornLUniversal Reference RNA is not a representative normal sample for oligonucleotide microarray studiesPathol Oncol Res20081432435118553159
- MojicaWDSteinLHawthornLAn exfoliation and enrichment strategy results in improved transcriptional profiles when compared to matched formalin fixed samplesBMC Clin Pathol20077717683544
- BurgemeisterRNucleic acids extraction from laser microdissected FFPE tissue sectionsMethods Mol Biol201172411712921370010
- ShiLPerkinsRGFangHTongWReproducible and reliable microarray results through quality control: Good laboratory proficiency and appropriate data analysis practices are essentialCurr Opin Biotechnol2008191101818155896
- ShiLReidLHJonesWDThe MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurementsNat Biotechnol20062491151116116964229
- NovoradovskayaNWhitfieldMLBasehoreLSUniversal reference RNA as a standard for microarray experimentsBMC Genomics2004512015113400
- AuerHLyianarachchiSNewsomDKlisovicMIMarcucciGKornackerKChipping away at the chip bias: RNA degradation in microarray analysisNat Genet200335429229314647279
- VandesompeleJDe PreterKPattynFAccurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genesGenome Biol200237 RESEARCH0034
- HellemansJMortierGDe PaepeASpelemanFVandesompeleJqBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR dataGenome Biol200782R1917291332
- BeckerCHammerle-FickingerARiedmaierIPfafflMWmRNA and microRNA quality control for RT-qPCR analysisMethods201050423724320079844
- DevonshireASElaswarapuRFoyCAEvaluation of external RNA controls for the standardisation of gene expression biomarker measurementsBMC Genomics20101166221106083
- BakerSCBauerSRBeyerRPThe external RNA controls consortium: A progress reportNat Methods200521073173416179916
- Invitrogen. Available from: www.invitrogen.com. Accessed May 7, 2011.
- VWR International Available from: https://ca.vwr.com. Accessed August 25, 2011.
- FanXFangHHongHPerkinsRShiLTongWCorrelation analysis of external RNA controls reveals its utility for assessment of microarray assayAnal Biochem2009385220320719059192
- WalterMHoneggerASchweizerRPothsSBoninMUtilization of AFFX spike-in control probes to monitor sample identity throughout Affymetrix GeneChip Array processingBiotechniques201048537137820569210
- StaalFJCarioGCazzanigaGConsensus guidelines for microarray gene expression analyses in leukemia from three European leukemia networksLeukemia20062081385139216761018
- RudloffUBhanotUGeraldWBiobanking of human pancreas cancer tissue: Impact of ex-vivo procurement times on RNA qualityAnn Surg Oncol20101782229223620162455
- BotlingJMickePBiobanking of fresh frozen tissue from clinical surgical specimens: Transport logistics, sample selection, and histologic characterizationMethods Mol Biol201167529930620949397
- BotlingJMickePFresh frozen tissue: RNA extraction and quality controlMethods Mol Biol201167540541320949406
- BotlingJEdlundKSegerstenUImpact of thawing on RNA integrity and gene expression analysis in fresh frozen tissueDiagn Mol Pathol2009181445219214109
- VermeulenJDerveauxSLefeverSRNA pre-amplification enables large-scale RT-qPCR gene-expression studies on limiting sample amountsBMC Res Notes2009223519930725
- Gonzalez-RocaEGarcia-AlbenizXRodriguez-MuleroSGomisRRKornackerKAuerHAccurate expression profiling of very small cell populationsPLoS One2010512e1441821203435
- CorbiFCInaokaRJFelixRSComparative expression of a set of genes to an internal housekeeping control in CDNA amplified and not amplified by PolyAPCR in non-Hodgkin’s lymphoma samples obtained from fine-needle aspiration cytologyDiagn Mol Pathol2010191404420186011
- FerreiraENMaschiettoMSilvaSDBrentaniHCarraroDMEvaluation of quantitative rt-PCR using nonamplified and amplified RNADiagn Mol Pathol2010191455320186012
- SpurgeonSLJonesRCRamakrishnanRHigh throughput gene expression measurement with real time PCR in a microfluidic dynamic arrayPLoS One200832e166218301740
- GlavesPDTugwoodJDGeneration and analysis of transcriptomics dataMethods Mol Biol201169116718520972753
- LippaKADuewerDLSalitMLGameLCaustonHCExploring the use of internal and external controls for assessing microarray technical performanceBMC Res Notes2010334921189145
- FengLLiuHLiuYPower of deep sequencing and agilent microarray for gene expression profiling studyMol Biotechnol201045210111020432071
- CanalesRDLuoYWilleyJCEvaluation of DNA microarray results with quantitative gene expression platformsNat Biotechnol20062491115112216964225
- CoppéeJYDo DNA microarrays have their future behind them?Microbes Infect20081091067107118662797
- Anonymous.Quality System (QS) Regulation/Medical Device Good Manufacturing PracticesSilver Spring, MDFood and Drug Administration2011 Available from: www.fda.gov/MedicalDevices/DeviceRegulationandGuidance/PostmarketRequirements/QualitySystemsRegulations. Accessed May 7, 2011.
- International Organization for StandardizationISO 900 EssentialsGeneva, Switzerland Available from: www.iso.org/iso/iso_9000_essentials. Accessed May 7, 2011.
- BengtssonMStåhlbergARorsmanPKubistaMGene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levelsGenome Res200515101388139216204192
- RitchieMESilverJOshlackAA comparison of background correction methods for two-colour microarraysBioinformatics200723202700270717720982
- DurbinBPRockeDMVariance-stabilizing transformations for two-color microarraysBioinformatics200420566066715033873
- FanJNiuYSelection and validation of normalization methods for c-DNA microarrays using within-array replicationsBioinformatics200723182391239817660210
- McMullenPDMorimotoRIAmaralLANPhysically grounded approach for estimating gene expression from microarray dataProc Natl Acad Sci U S A201010731136901369520643961
- ShiLCampbellGJonesWDThe MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive modelsNat Biotechnol201028882783820676074
- HackettJLArcherJKGaigalasAKLaboratory Standards Institute Document MM12-A: Diagnostic nucleic acid microarrays approved guidelineWayne, PAClinical and Laboratory Standards Institute2006 Available from: http://www.clsi.org/source/orders/free/mm12AF.pdf. Accessed August 18, 2011.
- HubbleJDemeterJJinHImplementation of GenePattern within the Stanford Microarray DatabaseNucleic Acids Res.200937Database issueD89890118953035
- SimonRAnalysis of DNA microarray expression dataBest Pract Res Clin Haematol200922227128219698933
- ParryRMJonesWStokesTHk-Nearest neighbor models for microarray gene expression analysis and clinical outcome predictionPharmacogenomics J201010429230920676068
- MonzonFAMedeirosFLyons-WeilerMHennerWDIdentification of tissue of origin in carcinoma of unknown primary with a microarray-based gene expression testDiagn Pathol20105320205775
- SchultzKRBowmanWPAledoAImproved early event-free survival with imatinib in Philadelphia chromosome-positive acute lymphoblastic leukemia: A Children’s Oncology Group studyJ Clin Oncol200927315175518119805687
- IzraeliSApplication of genomics for risk stratification of childhood acute lymphoblastic leukaemia: From bench to bedside?Br J Haematol2010151211913120678159
- JelierRGoemanJJHettneKMSchuemieMJden DunnenJTt’HoenPALiterature-aided interpretation of gene expression data with the weighted global testBrief Bioinform292011 [Epub ahead of print.]
- GudgeonJMMcClainMRPalomakiGEWilliamsMSRapid ACCE: Experience with a rapid and structured approach for evaluating gene-based testingGenet Med20079747347817666894
- GulleyMLBrazielRMHallingKCClinical laboratory reports in molecular pathologyArch Pathol Lab Med2007131685286317550311
- SchiffGDHasanOKimSDiagnostic error in medicine: Analysis of 583 physician-reported errorsArch Intern Med2009169201881188719901140
- SotiriouCPiccartMJTaking gene-expression profiling to the clinic: When will molecular signatures become relevant to patient care?Nat Rev Cancer20077754555317585334
- Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working GroupRecommendations from the EGAPP Working Group: Can tumor gene expression profiling improve outcomes in patients with breast cancer?Genet Med2009111667319125125
- MookSBonnefoiHPruneriGDaily clinical practice of fresh tumour tissue freezing and gene expression profiling: Logistics pilot study preceding the MINDACT trialEur J Cancer20094571201120819232484
- National Comprehensive Cancer NetworkNCCN Chronic Myelogenous Leukemia Clinical Practice Guidelines in Oncology (Version 2.2011)2011 Available from: www.nccn.org. Accessed June 7, 2011.
- HarrisLFritscheHMennelRAmerican Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancerJ Clin Oncol200725335287521217954709
- National Comprehensive Cancer NetworkNCCN Breast Cancer Clinical Practice Guidelines in Oncology (Version 2.2011) Available from: www.nccn.org. Accessed June 7, 2011.
- RiversPADobalianAGerminarioFAA review and analysis of the clinical laboratory improvement amendment of 1988: Compliance plans and enforcement policyHealth Care Manage Rev20053029310215923911
- United States Food and Drug AdministrationUnited States Food and Drug Administration MicroArray Quality Control (MAQC) project Available from: http://www.fda.gov/ScienceResearch/BioinformaticsTools/MicroarrayQualityControlProject/default.htm. Accessed June 7, 2011.
- FanXLobenhoferEKChenMConsistency of predictive signature genes and classifiers generated using different microarray platformsPharmacogenomics J201010424725720676064
- ShiWBessarabovaMDosymbekovDFunctional analysis of multiple genomic signatures demonstrates that classification algorithms choose phenotype-related genesPharmacogenomics J201010431032320676069
- TillinghastGWMicroarrays in the clinicNat Biotechnol201028881081220697405
- BustinSABeaulieuJFHuggettJMIQE precis: Practical implementation of minimum standard guidelines for fluorescence-based quantitative real-time PCR experimentsBMC Mol Biol2010117420858237
- College of American Pathologists Available from: www.cap.org. Accessed May 7, 2011.
- WarringtonJACorbisierPFeilotterHClinical Laboratory Standards Institute Document MM16-A: Use of external RNA controls in gene expression assays approved guidelineWayne, PAClinical and Laboratory Standards Institute2006
- WilsonJAZoccoliMAJacobsonJWClinical Laboratory Standards Institute Document MM17-A: Verification and validation of multiplex nucleic acid assays approved guidelineWayne, PAClinical and Laboratory Standards Institute2008
- ValdesRPayneDLinderMWGuidelines and recommendations for laboratory analysis and application of pharmacogenetics to clinical practiceWashington, DCNational Academy of Clinical Biochemistry2007
- SzecsiPBOdumLError tracking in a clinical biochemistry laboratoryClin Chem Lab Med200947101253125719663542
- FabbrettiGRisk management: Correct patient and specimen identification in a surgical pathology laboratory. The experience of Infermi Hospital, Rimini, ItalyPathologica201010239610121171512
- ValensteinPNAlpernGAKerenDFResponding to large-scale testing errorsAm J Clin Pathol2010133344044620154282
- StainesHGarcia-FernandezLPogothataRWallacePMacKayWVan LoonAMonitoring performance of nucleic acid-based diagnostic measurement system users by EQAAccreditation and Quality Assurance: Journal for Quality, Comparability and Reliability in Chemical Measurement2009145243252
- TholenDWBLBooneDJCooperWGClinical Laboratory Standards Institute document GP27-A2: Using proficiency testing to improve the clinical laboratory approved guideline2nd ed.Wayne, PAClinical and Laboratory Standards Institute2007
- MadejRMCaoZDolingerDLHallLNeuwaldPWilliamsLOClinical and Laboratory Standards Institute Document MM14-A: Proficiency testing (external quality assessment) for molecular methods approved guidelineWayne, PAClinical and Laboratory Standards Institute2005
- Quality Control for Molecular Diagnostics Available from: www.qcmd.org. Accessed May 7, 2011.
- SarewitzSJGeorgeHMillerWGTholenDWValensteinPNClinical Laboratory Standards Institute Document GP29-A2: Assessment of laboratory tests when proficiency testing is not available approved guideline2nd ed.Wayne, PAClinical and Laboratory Standards Institute2008
- Association for Molecular PathologyAMP Test Directory Available from: www.amptestdirectory.org. Accessed May 7, 2011.
- National Center for Biotechnology InformationGeneTests Available from: www.genetests.org. Accessed May 7, 2011.
- BildAHParkerJSGustafsonAMAn integration of complementary strategies for gene-expression analysis to reveal novel therapeutic opportunities for breast cancerBreast Cancer Res2009114R5519638211
- HuZFanCLivasyCA compact VEGF signature associated with distant metastases and poor outcomesBMC Med20097919291283
- HuangJShiWZhangJGenomic indicators in the blood predict drug-induced liver injuryPharmacogenomics J201010426727720676066
