Abstract
Genetic polymorphism for cytochrome 450 (P450) enzymes leads to interindividual variability in the plasma concentrations of many drugs. In some cases, P450 genotype results in decreased enzyme activity and an increased risk for adverse drug effects. For example, individuals with the CYP2D6 loss-of-function genotype are at increased risk for ventricular arrhythmia if treated with usual does of thioridazine. In other cases, P450 genotype may influence the dose of a drug required to achieve a desired effect. This is the case with warfarin, with lower doses often necessary in carriers of a variant CYP2C9*2 or *3 allele to avoid supratherapeutic anticoagulation. When a prodrug, such as clopidogrel or codeine, must undergo hepatic biotransformation to its active form, a loss-of-function P450 genotype leads to reduced concentrations of the active drug and decreased drug efficacy. In contrast, patients with multiple CYP2D6 gene copies are at risk for opioid-related toxicity if treated with usual doses of codeine-containing analgesics. At least 25 drugs contain information in their US Food and Drug Administration-approved labeling regarding P450 genotype. The CYP2C9, CYP2C19, and CYP2D6 genes are the P450 genes most often cited. To date, integration of P450 genetic information into clinical decision making is limited. However, some institutions are beginning to embrace routine P450 genotyping to assist in the treatment of their patients. Genotyping for P450 variants may carry less risk for discrimination compared with genotyping for disease-associated variants. As such, P450 genotyping is likely to lead the way in the clinical implementation of pharmacogenomics. This review discusses variability in the CYP2C9, CYP2C19, and CYP2D6 genes and the implications of this for drug efficacy and safety.
Introduction
It is well recognized that variation in the genes for cytochrome P450 (P450) enzymes contributes to interindividual differences in the plasma concentrations of drug substrates, resulting in interpatient variability in drug efficacy and safety. Functional polymorphism has been discovered for CYP2A6, CYP1A2, CYP2C9, CYP2C19, CYP2D6, and CYP3A4/5. At least 25 drugs now contain pharmacogenetic information related to P450 enzymes in their US Food and Drug Administration-approved labeling.Citation1 Examples of such drugs are provided in . The P450 genes included in drug labeling are limited to CYP2C9, CYP2C19, and CYP2D6. Thus, this review will focus on genetic variation for the CYP2C9, CYP2C19, and CYP2D6 enzymes and drug substrates for these enzymes.
Table 1 Examples of drugs with genotype information included in their US Food and Drug Administration-approved labelingCitation1
CYP2C9 genotype
Functionality of CYP2C9 variants
The CYP2C9 enzyme metabolizes approximately 15% of clinically used drugs, including some anticoagulants (eg, S-warfarin), hypoglycemics (eg, tolbutamide), angiotensin II receptor blockers (eg, losartan), antiepileptics (eg, phenytoin), and nonsteroidal anti-inflammatory drugs (eg, diclofenac).Citation2 To date, 35 variants of the CYP2C9 gene have been identified.Citation3 There are racial differences in the frequency of CYP2C9 variants, as shown in .Citation4–Citation6 The CYP2C9*2 allele is common in Caucasians and results from the R144C substitution located on the exterior surface of the enzyme.Citation7 This allele leads to decreased CYP2C9 enzyme activity in vitro, the magnitude of which ranges from 8% to 94% depending on the CYP2C9 substrate (reviewed in Lee et alCitation8). The CYP2C9*3 allele is also common in Caucasians and results from the I359L substitution at the substrate recognition site of the enzyme.Citation9 This leads to a 71%–96% decrease in enzyme activity (also reviewed in Lee et alCitation8). The decreased catalytic activities of CYP2C9*2 and *3 are in part due to enhanced uncoupling (ie, abortive catalytic cycle) of the reaction or a disruption of the water network in the variant enzymes.Citation10 The CYP2C9*5, *6, *8, and *11 alleles predominate in African populations, with decreased enzyme activity reported with the *5, *6, and *11 alleles.Citation11–Citation13 The CYP2C9*8 allele is the most common of these alleles in African Americans and results from the R150H substitution in exon 3. Data on the effects of the CYP2C9*8 allele on enzyme activity are inconsistent. Although an in vitro study showed increased tolbutamide metabolism with the CYP2C9*8 variant compared with the wild type,Citation13 in vivo studies using phenytoin or losartan revealed decreased or no change in drug elimination in CYP2C9*8 carriers.Citation12,Citation14 This discrepancy may reflect substrate-specific effects of CYP2C9 variants on enzyme activity, as has been previously reported for the CYP2C9*2 and *3 alleles,Citation8,Citation15 the underlying molecular mechanisms for which remain to be characterized.
Impact of CYP2C9 genotype on warfarin response
Warfarin is the most commonly prescribed oral agent for the prevention of thromboembolism. Warfarin has a narrow therapeutic index and is dosed according to the international normalized ratio (INR), with an INR of two to three recommended for most indications.Citation16 The risk for thrombosis increases with subtherapeutic anticoagulation,Citation17,Citation18 while the risk for bleeding increases significantly when the INR exceeds four.Citation19 Warfarin is a challenging drug to manage, largely because the dose required to achieve a therapeutic INR varies as much as 20-fold among individuals.Citation20
Warfarin is a racemic mixture, and the more potent S-enantiomer is metabolized almost exclusively by CYP2C9 to the inactive 7-hydroxy metabolite. The clearance of S-warfarin is reduced approximately 40% with the CYP2C9*1/*2 genotype, up to 75% with the *1/*3 genotype, and by as much as 90% with the *3/*3 genotype.Citation21,Citation22 Accordingly, lower warfarin doses are generally required in the presence of a CYP2C9*2 or *3 allele.Citation4,Citation5,Citation20,Citation23–Citation26 For example, Taube et alCitation27 reported that patients with a variant CYP2C9*2 or *3 allele required between 61% and 86% of the dose needed by *1 allele homozygotes.
shows warfarin dose requirements by CYP2C9 genotype across racial groups according to data from the International Warfarin Pharmacogenetics Consortium, a collaboration of 21 research groups from four continents who have pooled genotype and phenotype data from over 5700 warfarin-treated patients.Citation28,Citation29 As shown, CYP2C9 genotype influences warfarin dose requirements across racial groups. In genome-wide association studies in Caucasians, the CYP2C9*2 and *3 alleles were shown to explain 9%–12% of the total variability in warfarin dose requirements.Citation30,Citation31
Table 3 Median warfarin dose requirements by CYP2C9 genotype across racial groups, according to the data from the International Warfarin Pharmacogenetics ConsortiumCitation29
African Americans are underrepresented in warfarin pharmacogenomic studies. However, recent data show significantly lower warfarin dose requirements among patients with a CYP2C9*5, *6, *8, or *11 allele compared to those with the *1/*1 genotype.Citation5 Specifically, among 226 African Americans, the warfarin maintenance dose was 18% lower in individuals with a CYP2C9*5, *6, *8, or *11 allele compared to those with the *1/*1 genotype (median dose of 5.0 mg/day versus 6.1 mg/day, P = 0.004). The CYP2C9 genotype explained approximately 8% of the total variance in warfarin dose, just slightly less than that in Caucasians. In a recent targeted resequencing approach, Perera et alCitation32 found a novel CYP2C9 single nucleotide polymorphism in the African American genome that was associated with higher warfarin dose requirements. The 18786A > T single nucleotide polymorphism is located in intron three and occurs in 40% of African Americans. In a cohort of over 300 African Americans, investigators observed a 3.7 mg/week increase in warfarin dose requirements for each 18786T allele.
The CYP2C9 genotype has also been implicated as a risk factor for over-anticoagulation and bleeding with warfarin.Citation23,Citation24,Citation26,Citation33 Limdi et alCitation33 conducted one of the largest studies to examine bleeding risk according to CYP2C9 genotype. Among 446 patients started on warfarin, including 227 African Americans, investigators observed 44 major bleeding events during a 2-year follow-up period. The variant CYP2C9 *2, *3, *5, *6, and *11 alleles conferred an increased risk of major bleeding, with an adjusted hazard ratio of 3.0 (95% confidence interval of 1.2–7.5). The bleeding risk was similar between Caucasians and African Americans. Interestingly, while the association between CYP2C9 genotype and risk for bleeding was highest during warfarin initiation, it persisted after dose stabilization. These data suggest that bleeding should be closely monitored throughout warfarin therapy for patients with a variant CYP2C9 allele. Based on the totality of data, CYP2C9 variants appear to increase the bleeding risk with warfarin approximately twofold.Citation34
There are data suggesting that CYP2C9 gene expression changes during development, and this, in turn, may lead to age-related differences in the CYP2C9 genotype-drug response phenotype.Citation35 In support of differential genotype effects by age, a recent study showed minimal influence of CYP2C9 genotype on warfarin dose variability in pediatric patients.Citation36 Specifically, among 59 children (aged 1–19 years) treated with either warfarin or the vitamin K antagonist phenprocoumon, CYP2C9 genotype explained <1% of the variability in vitamin K antagonist dose. Age was the most important predictor of dose requirement, explaining 28% of variance in dose.
Two small prospective trials and a comparative effectiveness study provide evidence of clinical benefit with genotype-guided warfarin dosing in adult populations.Citation37–Citation39 Caraco et alCitation37 randomized 191 patients to warfarin dosing based on either clinical factors plus CYP2C9 genotype or clinical factors alone. Genotype-guided dosing resulted in more rapid attainment of stable anticoagulation, more time spent within the therapeutic range, and a lower incidence of minor bleeding than dosing according to clinical factors alone. Similarly, Huang et alCitation39 reported that dosing based on genotypes for CYP2C9 and vitamin K epoxide reductase complex 1 (VKORC1), which codes for the primary target of warfarin, improved the time to achieve stable dosing in a small Asian population.
Epstein et alCitation38 evaluated the effect of CYP2C9 and VKORC1 genotyping at the time of warfarin initiation on clinical outcomes. In this comparative effectiveness study, the incidence of hospitalization was compared between a group of 896 patients offered free genotyping and a control group of nearly 2700 patients who started warfarin the previous year and were not offered genotyping. Genotype results were provided to each patient’s physician with an interpretative report. Those who underwent genotyping had fewer hospitalizations for any cause and fewer hospitalizations for bleeding or thromboembolism during the 6-month follow-up period compared with controls.
Findings from these positive studies are tempered by findings from two small (n = 206–230) prospective trials of Caucasians showing no benefit with genotype-guided warfarin dosing in terms of time spent in the therapeutic INR range during the initial months of therapy.Citation40,Citation41 Several large multicenter randomized-controlled clinical trials are underway to further assess the clinical utility of warfarin pharmacogenetics. One of the largest of these is the National Heart, Lung, and Blood Institute-sponsored Clarification of Optimal Anticoagulation Through Genetics trial.Citation42 This trial is targeting an enrollment of 1238 patients randomized to either a genotype-guided or clinical warfarin-dosing strategy. The study will assess the primary outcome of percent of time spent within the therapeutic range and is expected to be completed in early 2012.
In 2007, warfarin labeling was revised to include pharmacogenetic data. The label was further revised in 2010 to include a dosing table based on CYP2C9 and VKORC1 genotypes.Citation43 At least four US Food and Drug Administration-cleared genotyping assays are available for clinical use. There are also a number of published algorithms to assist clinicians with warfarin dosing when CYP2C9 genotype is known.Citation28,Citation44–Citation46 However, a limitation of most genotyping assays and algorithms is that they do not include CYP2C9 variants that are common in African Americans and thus may have lower utility in this population. Many clinicians and third-party payers are awaiting results of ongoing clinical trials before embracing genotype-guided warfarin dosing.
CYP2C19 genotype
Functionality of CYP2C19 variants
The CYP2C19 enzyme metabolizes approximately 10% of clinically used drugs, including S-mephenytoin, proton pump inhibitors (PPIs), and nelfinavir. The CYP2C19 enzyme is also responsible for biotransformation of clopidogrel to its pharmacologically active form. Genetic deficiency in CYP2C19-mediated S-mephenytoin elimination was first reported in 1979,Citation47 with the urinary excretion rate of 4-hydroxymephenytoin (a metabolite of S-mephenytoin produced by CYP2C19)Citation48 used to discriminate extensive metabolizers (EMs; possessing normal enzyme activity) from poor metabolizers (PMs; with reduced or absent enzyme activity) for CYP2C19 substrates.Citation49,Citation50
To date, 28 CYP2C19 variants have been identified.Citation51 The CYP2C19*1 allele is considered the wild-type allele, with normal enzyme activity. The CYP2C19*2 allele is the primary allele responsible for the PM phenotype in Asians and Caucasians.Citation52 Other, less common, defective alleles leading to the PM phenotype include CYP2C19*3 in AsiansCitation53 and CYP2C19*4, *5, *6, *7, and *8 in Caucasians.Citation54 The CYP2C19*2 through *8 alleles are deemed loss-of-function alleles. Both CYP2C19*2 (c.681G>A) and *3 (c.636G>A) produce a truncated nonfunctional protein. The CYP2C19*2 allele results from a splicing defect in exon 5, while the CYP2C19*3 allele results from a single base substitution leading to a premature stop codon in exon 4.Citation52 Various base-pair sequence changes in the CYP2C19*4, *5, *6, *7, and *8 alleles negatively influence expression of the protein and catalytic activity.Citation55 However, the CYP2C19*17 allele results from polymorphisms in the gene promoter region (c.99C>T and c.991A>G) and increases enzyme activity due to enhanced gene transcription. Thus, CYP2C19*17 is deemed a gain-of-function allele.Citation56 CYP2C19 genotype confers four phenotypes: extensive metabolism (*1/*1), poor metabolism (eg, *2/*2, *2/*3, or *3/*3), intermediate metabolism (eg, *1/*2,), and ultrarapid metabolism (*1/*17, *17/*17). There are marked racial differences in the frequencies of CYP2C19 phenotypes, as shown in , with Asians having the highest frequencies of the PM and intermediate metabolizer (IM) phenotypes.Citation57,Citation58
Table 4 The frequency of CYP2C19 phenotypes by raceCitation53,Citation57,Citation58
Impact of CYP2C19 genotype on clopidogrel effectiveness
Clopidogrel is an antiplatelet agent that is widely used in patients with cardiovascular disease. Clopidogrel, in combination with aspirin, has been shown to reduce morbidity and mortality in patients with an acute coronary syndrome (ACS) who are either managed medically or with coronary revascularization.Citation59–Citation61 Dual antiplatelet therapy with clopidogrel plus aspirin also reduces the risk for coronary stent thrombosis following percutaneous coronary intervention (PCI).Citation62 There is marked interpatient variability in clopidogrel’s effectiveness, with approximately 25% of treated patients exhibiting residual ex vivo platelet aggregation.Citation63 These patients are at increased risk for major adverse cardiac events, including myocardial infarction and stent thrombosis.Citation64 The variability in clopidogrel response is largely attributed to interpatient differences in clopidogrel pharmacokinetics.
Clopidogrel requires hepatic bioactivation to its active thiol metabolite, which irreversibly binds to the platelet P2Y12 receptor, thus inhibiting platelet activation and subsequent aggregation. Approximately 85% of the oral dose is inactivated by esterases, leaving only 15% available for hepatic bioactivation to the active metabolite. Bioactivation of clopidogrel is a two-step oxidative process, as shown in . The CYP2C19 enzyme is involved in both oxidative steps. Reduced or absent CYP2C19 activity, secondary to either genetic polymorphism or interacting medication (eg, PPIs) results in decreased exposure to the active thiol metabolite, which, in turn, diminishes clopidogrel’s effectiveness.
Figure 1 Bioactivation and mechanism of action of clopidogrel, prasugrel, and ticagrelor. CYP2C19 (in bold) is the predominant enzyme in clopidogrel bioactivation. The P2Y12 receptor on the surface of the platelet is the site of action of clopidogrel, prasugrel, and ticagrelor.
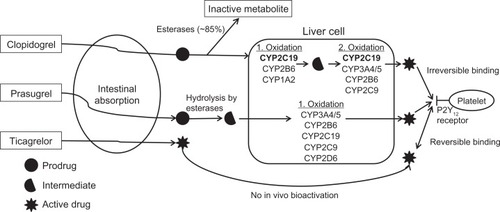
IMs and PMs produce approximately 30%–50% less of the active clopidogrel metabolite compared with EMs.Citation65–Citation68 As a result, IMs and PMs may derive less protection from thrombotic events with clopidogrel. Genetic substudies of a number of large clinical trials have demonstrated a higher incidence of stent thrombosis and major adverse cardiovascular events (ie, cardiovascular death, myocardial infarction, or stroke) following ACS or PCI among clopidogrel-treated patients with the PM or IM phenotype compared with similarly treated EMs.Citation68–Citation76
Mega et alCitation77 conducted a meta-analysis of nine clinical trials including 9685 patients, of whom 54% had ACS and 91% underwent PCI. The hazard ratio for major adverse cardiovascular events was 1.55 (95% confidence interval 1.11–2.17) in carriers of one loss-of-function allele (IMs) and 1.76 (95% confidence interval 1.24–2.50) for carriers of two loss-of-function alleles (PMs), compared with noncarriers. Among patients who underwent PCI with stent placement, the hazard ratio for stent thrombosis was 2.67 (95% confidence interval 1.69–4.22) and 3.97 (95% confidence interval 1.75–9.02) in carriers of one or two loss-of-function alleles, respectively, compared with noncarriers.
In contrast, a substudy of two large placebo-controlled trials showed no effect of CYP2C19 loss-of-function genotype on clopidogrel efficacy in the setting of ACS or atrial fibrillation.Citation78 However, there are two caveats with this substudy that are worth mentioning. First, fewer than 15% of patients with ACS underwent stent placement and none had ST segment elevation ACS. Secondly, the benefits of clopidogrel in the setting of atrial fibrillation are minimal.Citation79 Thus, while these data suggest that reduced CYP2C19 activity does not impact clopidogrel efficacy in patients with lower-risk ACS who are predominately medically managed, an insufficient number of patients who underwent coronary revascularization were included to reach any conclusions in this higher-risk group.
There is some evidence suggesting an increased risk of bleeding with clopidogrel in ultrarapid metabolizers (UMs) with the CYP2C19*17 allele. Specifically, among over 1500 patients who underwent PCI and stent placement, carriers of a CYP2C19*17 allele had an increased risk for both major and minor bleeding with clopidogrel compared with noncarriers.Citation58 The risk for bleeding was greatest among *17 homozygotes, with an odds ratio of 3.27 (95% confidence interval 1.33–8.10) compared with noncarriers of the *17 allele.
The clopidogrel labeling was updated in March 2010 in response to reports of reduced efficacy with CYP2C19 loss-of-function alleles.Citation80 The label warns of reduced effectiveness in PMs and states that genetic testing for reduced-function CYP2C19 alleles is available. The label further advises health care professionals to consider alternative strategies in patients identified as PMs. However, the labeling falls short of providing recommendations on whom to genotype and what specific approaches to undertake in patients testing positive for a loss-of-function allele. Additionally, the labeling does not address IMs, who are clearly at increased risk for adverse cardiovascular events compared with EMs, although at lower risk than PMs.
Recently, the National Institutes of Health-supported Clinical Pharmacogenetics Implementation Consortium published guidelines on genotype-guided antiplatelet therapy in cardiovascular disease.Citation81 These guidelines do not make firm recommendations about which patients should be genotyped but instead suggest two potential approaches. The first approach is to genotype all patients with ACS or undergoing PCI; the second is to target moderate-to-high-risk patients, such as those with a history of stent thrombosis, diabetes, renal insufficiency, or high-risk coronary angiographic features. In patients who have CYP2C19 genotype data available, standard dose clopidogrel is recommended for EMs and UMs. Alternative therapy with prasugrel, ticagrelor, or cilostazol is recommended for IMs or PMs.
Similar to clopidogrel, prasugrel is a thienopyridine that irreversibly binds to the platelet P2Y12 receptor and requires hepatic bioactivation (). While CYP2C19 is involved in prasugrel bioactivation, CYP2C19 genotype does not affect the generation of active metabolite or drug efficacy, probably because the reaction is not highly dependent on CYP2C19.Citation66,Citation67,Citation82,Citation83 Ticagrelor is a recently approved reversible P2Y12 antagonist that does not require hepatic bioactivation. As such, ticagrelor pharmacokinetics is not affected by CYP2C19 genotype.Citation84 Cilostazol is a phosphodiesterase type III inhibitor shown to more effectively inhibit platelet aggregation than high-dose clopidogrel (eg, 150 mg/day) in CYP2C19 IMs and PMs.Citation85–Citation87 Use of high-dose clopidogrel (150 mg daily) is not recommended as an alternative therapy in PMs, based on recent data demonstrating no benefit with higher doses in patients with residual, ex vivo, platelet reactivity after PCI.Citation81,Citation88
Impact of CYP2C19 genotype on the efficacy of PPIs
PPIs, including omeprazole, esomeprazole, pantoprazole, lansoprazole, and rabeprazole, are partially metabolized by CYP2C19. Loss-of-function CYP2C19 alleles result in higher plasma concentrations of PPIs and greater suppression of gastric acid.Citation89–Citation91 Consistent with these data, higher Helicobacter pylori eradication rates are achieved with both dual (PPI and amoxicillin) and triple (PPI, amoxicillin, and clarithromycin) therapy in patients with a defective CYP2C19 allele. Specifically, reported cure rates with standard dose omeprazole (20 mg/day) plus amoxicillin were 100%, 60%, and 29%, in PMs, IMs, and EMs, respectively.Citation92 Rabeprazole 10 mg twice daily plus amoxicillin provided better H. pylori cure rates; although differences by genotype were observed (94%, 92%, and 61% in PMs, IMs, and EMs, respectively).Citation93 Using a higher dose of rabeprazole (10 mg four times daily) effectively eradicated H. pylori for EMs who failed initial therapy.Citation93 Triple therapy that included omeprazole 20 mg or lansoprazole 30 mg twice daily produced cure rates of 98%–100% for PMs but only 73%–86% in EMs.Citation94,Citation95 Of EMs who failed triple therapy, a 97% H. pylori eradication was achieved with high-dose lansoprazole (30 mg four times daily) and amoxicillin.Citation94
CYP2C19 genotype also impacts the effectiveness of PPIs for gastroesophageal reflux disease. Among patients with gastroesophageal reflux disease treated with lansoprazole 30 mg daily for 8 weeks, cure rates of mucosal breaks were 85% in PMs, 60% in IMs, and 46% in EMs.Citation96 Those with the EM phenotype and the most erosive esophagitis achieved cure rates of only 17%.
Similar to CYP2C9, there is evidence that CYP2C19 gene expression changes with development, with adult expression levels reached by age 10.Citation35 As a consequence, the CYP2C19 genotype-PPI response relationship may differ between adult and pediatric patients. In support of this, a single-dose pharmacokinetic study in children showed no effect of CYP2C19 genotype on omeprazole pharmacokinetics.Citation97 In contrast, CYP2C19 genotype clearly impacts omeprazole levels in adults.Citation91
Information regarding CYP2C19 genotype is included in the drug interaction and clinical pharmacology sections of rabeprazole and esomeprazole labels.Citation1 The data suggest that use of higher PPI doses will overcome reduced effectiveness in EMs.Citation93,Citation94 Dosing strategies for PPIs based on CYP2C19 genotype have been proposed for rabeprazole: 20 mg/day for PMs, 20 mg twice daily for IMs, and 10 mg four times daily for EMs.Citation98
Impact of CYP2C19 genotype on nelfinavir response in human immunodeficiency virus (HIV)
Nelfinavir is metabolized by CYP2C19 to the hydroxyl-tertbutylamide (M8) metabolite. CYP2C19 genotype can influence the bioavailability of nelfinavir, with lower oral clearance with the *2 allele.Citation99 Saitoh et alCitation99 examined the effect of CYP2C19 genotype on antiviral response to nelfinavir in 152 children taking highly active antiretroviral therapy. At 24 weeks, presence of the CYP2C19*2 allele was associated with a greater likelihood of achieving plasma HIV RNA < 400 copies/mL; 46%, 69%, and 63% of patients with the *1/*1, *1/*2, and *2/*2 genotypes, respectively, achieved concentrations <400 copies/mL. Others have reported a trend toward decreased virologic failure with the CYP2C19*2 allele.Citation100 Information regarding CYP2C19 genotype is included in the drug interaction and clinical pharmacology sections of the nelfinavir label. However, no recommendations are provided regarding genetic testing.
CYP2D6 genotype
Discussion of clinically relevant variants
The CYP2D6 enzyme metabolizes about 25% of clinically used drugs from many different drug classes including anti-depressants, antipsychotics, antihypertensives, and analgesics. Since early reports of a population exhibiting defective metabolism of debrisoquine or sparteine (CYP2D6 substrates) as a result of genotype,Citation101,Citation102 genetic polymorphisms leading to different CYP2D6 phenotypes have been extensively studied. Based on the extent of CYP2D6 substrate metabolism, the population is largely divided into four different groups, listed in order of decreasing CYP2D6 activity: UMs, EMs, IMs, and PMs. EMs carry at least one CYP2D6 allele showing normal enzyme function, IMs carry one CYP2D6 allele showing reduced function and another with no CYP2D6 function, and PMs carry two nonfunctional CYP2D6 alleles.Citation103 UMs, however, carry multiple copies of functional CYP2D6 genes.Citation104 The CYP2D6*1, *2, and *4 alleles are the most common alleles associated with the gene duplication and multiduplication. There are racial differences in the frequency of CYP2D6 alleles (). Ethiopian and Saudi Arabian populations have an unusually high (10%–16%) frequency of CYP2D6 gene duplication when compared with populations in other African or Asian countries, the cause of which has not been elucidated.Citation105
Table 5 Major CYP2D6 variants
Impact of CYP2D6 genotype on response to opioid analgesics
Codeine and tramadol are prodrugs requiring conversion via CYP2D6 to their therapeutically active metabolites, morphine and O-desmethyltramadol, respectively. Poor CYP2D6 metabolizers achieve lower metabolite concentrations and experience minimal analgesic relief with usual doses of these drugs.Citation106–Citation109 In contrast, patients with CYP2D6 gene duplication and the UM phenotype are at risk for toxic plasma concentrations of the morphine and O-desmethyltramadol metabolites after taking codeine or tramadol, respectively.Citation110–Citation112 Indeed, there are reports of severe respiratory depression and abdominal pain in patients treated with codeine-containing analgesics, who were subsequently found to have the UM phenotype.Citation111,Citation112 In addition, serious opioid-related toxicity and even death have been reported in breastfed infants of mothers with the UM phenotype.Citation113 These data suggest that codeine- and tramadol-containing analgesics should be reserved for patients with the EM phenotype to avoid analgesic failure in PMs and toxicity in UMs.
Impact of CYP2D6 genotype on response to antidepressants and antipsychotics
The CYP2D6 enzyme is involved in the metabolism of secondary (eg, protriptyline, nortriptyline) and tertiary (eg, doxepine) tricyclic antidepressants. The PM phenotype has been correlated with greater tricyclic antidepressant plasma concentrations and lower dose requirements.Citation114,Citation115 In contrast, extremely low nortriptyline concentrations have been observed in patients with multiple CYP2D6 gene copies.Citation116 Tricyclic antidepressant doses two to five times higher than normally recommended may be necessary to achieve therapeutic plasma concentrations in UMs.Citation117,Citation118 Pharmacogenetic information is included in the labeling of doxepine and protriptyline (). Lower doses of these drugs should be considered in patients with a reduced-function CYP2D6 allele.Citation1
The CYP2D6 genotype has also been associated with plasma concentrations of various selective serotonin reuptake inhibitors (SSRIs), such as paroxetine and fluoxetine.Citation119 For example, Charlier et alCitation120 reported significantly higher steady-state plasma concentrations of paroxetine and fluoxetine among PMs compared with EMs. However, results from various pharmacokinetic studies are inconsistent, and there is no clear correlation between CYP2D6 genotype and clinical response.Citation119 In a systematic review of 37 research articles, Thakur et alCitation119 concluded that data are insufficient to support CYP2D6 genotyping to guide SSRI prescribing.
Evidence to support CYP2D6 genotyping to predict efficacy of antipsychotic therapy is also limited.Citation121 However, there are data supporting an association between CYP2D6 polymorphisms and antipsychotic toxicity. The CYP2D6 enzyme extensively metabolizes the typical (eg, haloperidol, perphenazine, and thioridazine) and atypical (eg, risperidone, clozapine, and aripiprazole) antipsychotics. Higher plasma concentrations of antipsychotics, including perphenazine, haloperidol, thioridazine, and risperidone, are observed in individuals with reduced-function CYP2D6 alleles compared with EMs.Citation122,Citation123 In a meta-analysis of prospectively designed studies, Fleeman et alCitation121 reported that antipsychotic-treated patients with a nonfunctional CYP2D6 allele were at higher risk for tardive dyskinesia (odds ratio of 1.83, 95% confidence interval 1.09–3.08) and parkinsonism (odds ratio of 1.64, 95% confidence interval 1.04–2.58) compared with EMs.Citation121 However, the authors concluded that the differences observed by genotype were small and probably not of clinical significance.
Pharmacogenetic information is included in the labeling of thioridazine, clozapine, and aripiprazole. The language is strongest for thioridazine, which has the potential to prolong the QT interval in a dose-dependent manner, thus increasing the risk for life-threatening ventricular arrhythmias. The CYP2D6 PM phenotype is correlated with greater QT interval prolongation during thioridazine therapy.Citation124 As such, thioridazine is contraindicated in patients known to have reduced CYP2D6 activity. Recommendations for other antipsychotics are shown in .
Impact of CYP2D6 genotype on response to β-blockers
Beta-blockers are metabolized by CYP2D6, and CYP2D6 polymorphisms have significant effects on β-blocker plasma concentrations.Citation125,Citation126 However, data regarding the clinical significance of CYP2D6 genotype on β-blocker response are inconsistent. For example, Rau et alCitation125 reported significantly higher metoprolol plasma concentrations in PMs versus EMs (70 ng/mL versus 14 ng/mL), despite similar drug doses. This translated into greater reductions in heart rate and diastolic blood pressure in PMs. In contrast, Terra et alCitation126 found that, while S-metoprolol plasma concentrations differed significantly by CYP2D6 genotype among heart failure patients, genotype had no effect on β-blocker tolerability, heart rate, or systolic blood pressure response. Beta-blockers have a wide therapeutic index and thus genes affecting drug disposition may have minimal effects on drug response. Genes for drug target proteins may be more likely to be of clinical significance for β-blockers and other drugs with a wide therapeutic index. Indeed, polymorphisms in the gene encoding for the β1-adrenergic receptor are associated with clinical response to β-blockers in both hypertension and heart failure.Citation127,Citation128
Impact of CYP2D6 genotype on outcomes with tamoxifen therapy
Tamoxifen is widely prescribed for estrogen receptor-positive breast cancer. Tamoxifen requires biotransformation to its active 4-hydroxytamoxifen and endoxifen metabolites, which possess potent antiestrogenic effects. The CYP2D6 enzyme is a key enzyme in tamoxifen bioactivation. Tamoxifen metabolite levels are inversely correlated with the number of defective CYP2D6 alleles.Citation129,Citation130 The CYP2D6 genotype is estimated to explain 39% and 9% of the variability in endoxifen and 4- hydroxytamoxifen plasma concentrations, respectively.Citation129
The ability to metabolize tamoxifen has been associated with treatment outcomes.Citation131 In over 1300 patients with estrogen receptor-positive breast cancer, treatment with tamoxifen resulted in greater disease recurrence in patients with a reduced or loss-of-function CYP2D6 allele compared with those with normal enzyme activity. The highest recurrence rates occurred in patients with two loss-of-function (*3, *4, or *5) alleles (PMs), who had a nearly twofold higher incidence of recurrence (29% in PMs versus 14.9% in EMs). Similarly, patients with a reduced-function allele had worse event-free survival (hazard ratio 1.33, 95% confidence interval 1.06–1.68) and disease-free survival (hazard ratio 1.29, 95% confidence interval 1.03–1.61) than EMs. Other studies have shown similarly deleterious effects of CYP2D6 variant alleles on clinical outcomes with tamoxifen.Citation132–Citation134 However, the data are inconsistent and, in one case, conflicting.Citation135,Citation136 The discrepancies in the data may be secondary to factors other than CYP2D6 genotype influencing plasma concentrations of tamoxifen metabolites.Citation130 While data exist to support genotype-guided adjuvant endocrine therapy, the clinical utility of such an approach is currently unclear.Citation137,Citation138
Clinical relevance of P450 polymorphism
The potential consequences of P450 polymorphisms range from serious toxicity to ineffective drug therapy. Genetically determined reductions in P450 enzyme activity may have important implications for narrow-therapeutic-index drugs such as warfarin, where increased plasma concentrations contribute to drug toxicity. For prodrugs, such as codeine and clopidogrel, deficient enzyme activity can prevent the attainment of therapeutic drug plasma concentrations and lead to treatment failure. However, CYP2D6 gene duplication can lead to toxic reactions with codeine due to accumulation of the active morphine metabolite. For drugs with a wide therapeutic index, such as SSRIs and β-blockers, the clinical implications of P450 gene variation are less significant.
To date, clinicians have been slow to embrace pharmacogenetics, despite the addition of pharmacogenetic information in the labeling for many drugs. Two notable exceptions include clopidogrel and opioid analgesics. Vanderbilt University Medical Center, Nashville, TN, recently announced efforts to genotype for CYP2C19 variants in all patients with a high likelihood of requiring potent antiplatelet therapy in the future.Citation139 Genotype results are placed in the electronic medical record so that they may be used to assist in choosing appropriate antiplatelet therapy in the event of an ACS or PCI. Similarly, clinicians at St Jude Children’s Research Hospital, Memphis, TN, test patients for CYP2D6 genotype to individualize analgesic therapy.Citation140
Genotypes for the CYP2C9, CYP2C19, and CYP2D6 enzymes have minimal implications for disease susceptibility, as opposed to drug response. Specifically, an inherited deficiency in CYP2C9 may go completely undetected during an individual’s lifetime unless that individual is prescribed a narrow therapeutic index drug, such as warfarin, that relies on CYP2C9 for metabolism. Similarly, a person with inactive CYP2D6 may suffer no untoward effects unless exposed to a drug such as thioridazine that can produce deleterious cardiac consequences in CYP2D6 PMs. Thus, P450 genotyping may pose less ethical concerns than genotyping for disease-associated variants. As such, P450 genotyping is likely to lead the way in the clinical implementation of pharmacogenomics, as evidenced by current efforts at Vanderbilt University and St Jude Children’s Hospital. Genotyping arrays that broadly detect variation in multiple P450 genes are commercially available. Given the breadth of drugs that require P450 metabolism, use of such arrays may prove to be a feasible approach to broad implementation of P450 genotype-guided drug therapy.
Acknowledgements
This work was supported by the American Heart Association, Midwest Affiliate (10GRNT3750024) and National Institutes of Health, National Heart Lung and Blood Institute (HL106097).
Disclosure
Dr Cavallari is co-inventor of “CYP2C9*8 alleles correlate with decreased warfarin metabolism and increased warfarin sensitivity,” US Utility Patent Application No 12/572,908, published May 27, 2010; Pub No US 2010/0130599. The authors declare no other conflicts of interest in this work.
References
- US Food and Drug AdministrationTable of pharmacogenomic biomarkers in drug labels [web page on the Internet]Silver Spring, MDUS Food and Drug Administration2011 [updated September 27]. Available from: http://www.fda.gov/Drugs/ScienceResearch/ResearchAreas/Pharmacogenetics/ucm083378.htm. Accessed September 29, 2011
- MoSLZhouZWYangLPWeiMQZhouSFNew insights into the structural features and functional relevance of human cytochrome P450 2C9. Part ICurr Drug Metab200910101075112620167001
- SimSCCYP2CP allele nomenclature. Home page of the Human Cytochrome P450 (CYP) Allele Nomenclature Committee [website on the Internet] Human Cytochrome P450 (CYP) Allele Nomenclature Committee; 2011 [updated May 2]. Available from: http://www.cypalleles.ki.se/cyp2c9.htm. Accessed September 28, 2011.
- GageBFEbyCJohnsonJAUse of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarinClin Pharmacol Ther200884332633118305455
- CavallariLHLangaeeTYMomaryKMGenetic and clinical predictors of warfarin dose requirements in African AmericansClin Pharmacol Ther201087445946420072124
- TakahashiHWilkinsonGRNutescuEADifferent contributions of polymorphisms in VKORC1 and CYP2C9 to intra- and inter-population differences in maintenance dose of warfarin in Japanese, Caucasians and African-AmericansPharmacogenet Genomics200616210111016424822
- RettieAEWienkersLCGonzalezFJTragerWFKorzekwaKRImpaired (S)-warfarin metabolism catalysed by the R144C allelic variant of CYP2C9Pharmacogenetics19944139428004131
- LeeCRGoldsteinJAPieperJACytochrome P450 2C9 polymorphisms: a comprehensive review of the in-vitro and human dataPharmacogenetics200212325126311927841
- Sullivan-KloseTHGhanayemBIBellDAThe role of the CYP2C9-Leu359 allelic variant in the tolbutamide polymorphismPharmacogenetics1996643413498873220
- WeiLLocusonCWTracyTSPolymorphic variants of CYP2C9: mechanisms involved in reduced catalytic activityMol Pharmacol20077251280128817686967
- DickmannLJRettieAEKnellerMBIdentification and functional characterization of a new CYP2C9 variant (CYP2C9*5) expressed among African AmericansMol Pharmacol200160238238711455026
- AllabiACGalaJLHorsmansYCYP2C9, CYP2C19, ABCB1 (MDR1) genetic polymorphisms and phenytoin metabolism in a Black Beninese populationPharmacogenet Genomics2005151177978616220110
- BlaisdellJJorge-NebertLFCoulterSDiscovery of new potentially defective alleles of human CYP2C9Pharmacogenetics200414852753715284535
- AllabiACGalaJLHorsmansYFunctional impact of CYP2C95, CYP2C96, CYP2C98, and CYP2C911 in vivo among black AfricansClin Pharmacol Ther200476211311815289788
- TakanashiKTainakaHKobayashiKYasumoriTHosakawaMChibaKCYP2C9 Ile359 and Leu359 variants: enzyme kinetic study with seven substratesPharmacogenetics20001029510410761997
- AnsellJHirshJHylekEPharmacology and management of the vitamin K antagonists: American College of Chest Physicians Evidence-Based Clinical Practice Guidelines (8th Edition)Chest20081336 SupplS160S198
- HylekEMGoASChangYEffect of intensity of oral anticoagulation on stroke severity and mortality in atrial fibrillationN Engl J Med2003349111019102612968085
- KearonCGinsbergJSKovacsMJComparison of low-intensity warfarin therapy with conventional-intensity warfarin therapy for long-term prevention of recurrent venous thromboembolismN Engl J Med2003349763163912917299
- HylekEMEvans-MolinaCSheaCHenaultLEReganSMajor hemorrhage and tolerability of warfarin in the first year of therapy among elderly patients with atrial fibrillationCirculation2007115212689269617515465
- WadeliusMChenLYLindhJDThe largest prospective warfarin-treated cohort supports genetic forecastingBlood2009113478479218574025
- ScordoMGPengoVSpinaEDahlMLGusellaMPadriniRInfluence of CYP2C9 and CYP2C19 genetic polymorphisms on warfarin maintenance dose and metabolic clearanceClin Pharmacol Ther200272670271012496751
- TakahashiHKashimaTNomizoYMetabolism of warfarin enantiomers in Japanese patients with heart disease having different CYP2C9 and CYP2C19 genotypesClin Pharmacol Ther19986355195289630825
- AithalGPDayCPKestevenPJDalyAKAssociation of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complicationsLancet1999353915471771910073515
- HigashiMKVeenstraDLKondoLMAssociation between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapyJAMA2002287131690169811926893
- LimdiNABeasleyTMCrowleyMRVKORC1 polymorphisms, haplotypes and haplotype groups on warfarin dose among African-Americans and European-AmericansPharmacogenomics20089101445145818855533
- MargaglioneMColaizzoDD’AndreaGGenetic modulation of oral anticoagulation with warfarinThromb Haemost200084577577811127854
- TaubeJHalsallDBaglinTInfluence of cytochrome P-450 CYP2C9 polymorphisms on warfarin sensitivity and risk of over-anticoagulation in patients on long-term treatmentBlood20009651816181910961881
- KleinTEAltmanRBErikssonNEstimation of the warfarin dose with clinical and pharmacogenetic dataN Engl J Med2009360875376419228618
- LimdiNAWadeliusMCavallariLWarfarin pharmacogenetics: a single VKORC1 polymorphism is predictive of dose across 3 racial groupsBlood2010115183827383420203262
- CooperGMJohnsonJALangaeeTYA genome-wide scan for common genetic variants with a large influence on warfarin maintenance doseBlood200811241022102718535201
- TakeuchiFMcGinnisRBourgeoisSA genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dosePLoS Genet200953e1000433 Epub March 20, 200919300499
- PereraMAGamazonECavallariLHThe missing association: sequencing-based discovery of novel SNPs in VKORC1 and CYP2C9 that affect warfarin dose in African AmericansClin Pharmacol Ther201189340841521270790
- LimdiNAMcGwinGGoldsteinJAInfluence of CYP2C9 and VKORC1 1173C/T genotype on the risk of hemorrhagic complications in African-American and European-American patients on warfarinClin Pharmacol Ther200883231232117653141
- SandersonSEmeryJHigginsJCYP2C9 gene variants, drug dose, and bleeding risk in warfarin-treated patients: a HuGEnet systematic review and meta-analysisGenet Med2005729710415714076
- KoukouritakiSBManroJRMarshSADevelopmental expression of human hepatic CYP2C9 and CYP2C19J Pharmacol Exp Ther2004308396597414634042
- Nowak-GöttlUDietrichKSchaffranekDIn pediatric patients, age has more impact on dosing of vitamin K antagonists than VKORC1 or CYP2C9 genotypesBlood2010116266101610520833980
- CaracoYBlotnickSMuszkatMCYP2C9 genotype-guided warfarin prescribing enhances the efficacy and safety of anticoagulation: a prospective randomized controlled studyClin Pharmacol Ther200883346047017851566
- EpsteinRSMoyerTPAubertREWarfarin genotyping reduces hospitalization rates results from the MM-WES (Medco-Mayo Warfarin Effectiveness study)J Am Coll Cardiol201055252804281220381283
- HuangSWChenHSWangXQValidation of VKORC1 and CYP2C9 genotypes on interindividual warfarin maintenance dose: a prospective study in Chinese patientsPharmacogenet Genomics200919322623419177029
- AndersonJLHorneBDStevensSMRandomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulationCirculation2007116222563257017989110
- BurmesterJKBergRLYaleSHA randomized controlled trial of genotype-based Coumadin initiationGenet Med201113650951821423021
- FrenchBJooJGellerNLStatistical design of personalized medicine interventions: the Clarification of Optimal Anticoagulation through Genetics (COAG) trialTrials20101110821083927
- Coumadin® (warfarin sodium) [package insert]Princeton, NJBristol-Myers Squibb2010
- ShinJCaoDComparison of warfarin pharmacogenetic dosing algorithms in a racially diverse large cohortPharmacogenomics201112112513421174627
- WuAHWangPSmithADosing algorithm for warfarin using CYP2C9 and VKORC1 genotyping from a multi-ethnic population: comparison with other equationsPharmacogenomics20089216917818370846
- SconceEAKhanTIWynneHAThe impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimenBlood200510672329233315947090
- KüpferADesmondPVSchenkerSFamily study of a genetically determined deficiency of mephenytoin hydroxylation in man [letter]Pharmacologist197921173
- GoldsteinJAFalettoMBRomkes-SparksMEvidence that CYP2C19 is the major (S)-mephenytoin 4′-hydroxylase in humansBiochemistry1994337174317528110777
- WedlundPJAslanianWSMcAllisterCBWilkinsonGRBranchRAMephenytoin hydroxylation deficiency in Caucasians: frequency of a new oxidative drug metabolism polymorphismClin Pharmacol Ther19843667737806499356
- KüpferAPreisigRPharmacogenetics of mephenytoin: a new drug hydroxylation polymorphism in manEur J Clin Pharmacol19842667537596489416
- SimSCCYP2C9 allele nomenclature. Home page of the Human Cytochrome P450 (CYP) Allele Nomenclature Committee [website on the Internet] Human Cytochrome P450 (CYP) Allele Nomenclature Committee; 2011 [updated May 2]. Available from: http://www.cypalleles.ki.se/cyp2c19.htm. Accessed September 29, 2011
- de MoraisSWilkinsonGRBlaisdellJNakamuraKMeyerUAGoldsteinJAThe major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humansJ Biol Chem19942692215419154228195181
- GoldsteinJAIshizakiTChibaKFrequencies of the defective CYP2C19 alleles responsible for the mephenytoin poor metabolizer phenotype in various Oriental, Caucasian, Saudi Arabian and American black populationsPharmacogenetics19977159649110363
- WedlundPJThe CYP2C19 enzyme polymorphismPharmacology200061317418310971203
- DestaZZhaoXShinJGFlockhartDAClinical significance of the cytochrome P450 2C19 genetic polymorphismClin Pharmacokinet2002411291395812222994
- SimSCRisingerCDahlMLA common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressantsClin Pharmacol Ther200679110311316413245
- MomaryKMDorschMPFactors associated with clopidogrel nonresponsivenessFuture Cardiol20106219521020230261
- SibbingDKochWGebhardDCytochrome 2C19*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placementCirculation2010121451251820083681
- PatronoCBaigentCHirshJRothGAmerican College of Chest PhysiciansAntiplatelet drugs: American College of Chest Physicians Evidence-Based Clinical Practice Guidelines (8th Edition)Chest20081336 SupplS199S233
- ChenZMJiangLXChenYPAddition of clopidogrel to aspirin in 45,852 patients with acute myocardial infarction: randomised placebo-controlled trialLancet200536694971607162116271642
- YusufSZhaoFMehtaSREffects of clopidogrel in addition to aspirin in patients with acute coronary syndromes without ST-segment elevationN Engl J Med2001345749450211519503
- SteinhublSRBergerPBMannJT3rdEarly and sustained dual oral antiplatelet therapy following percutaneous coronary intervention: a randomized controlled trialJAMA2002288192411242012435254
- CombescureCFontanaPMalloukNClinical implications of clopidogrel non-response in cardiovascular patients: a systematic review and meta-analysisJ Thromb Haemost20108592393320156305
- BonelloLTantryUSMarcucciRConsensus and future directions on the definition of high on-treatment platelet reactivity to adenosine diphosphateJ Am Coll Cardiol2010561291993320828644
- UmemuraKFurutaTKondoKThe common gene variants of CYP2C19 affect pharmacokinetics and pharmacodynamics in an active metabolite of clopidogrel in healthy subjectsJ Thromb Haemost2008681439144118532997
- VarenhorstCJamesSErlingeDGenetic variation of CYP2C19 affects both pharmacokinetic and pharmacodynamic responses to clopidogrel but not prasugrel in aspirin-treated patients with coronary artery diseaseEur Heart J200930141744175219429918
- BrandtJTCloseSLIturriaSJCommon polymorphisms of CYP2C19 and CYP2C9 affect the pharmacokinetic and pharmacodynamic response to clopidogrel but not prasugrelJ Thromb Haemost20075122429243617900275
- MegaJLCloseSLWiviottSDCytochrome p-450 polymorphisms and response to clopidogrelN Engl J Med2009360435436219106084
- TrenkDHochholzerWFrommMFCytochrome P450 2C19 681G>A polymorphism and high on-clopidogrel platelet reactivity associated with adverse 1-year clinical outcome of elective percutaneous coronary intervention with drug-eluting or bare-metal stentsJ Am Coll Cardiol200851201925193418482659
- ColletJPHulotJSPenaACytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: a cohort studyLancet2009373966030931719108880
- GiustiBGoriAMMarcucciRRelation of cytochrome P450 2C19 loss-of-function polymorphism to occurrence of drug-eluting coronary stent thrombosisAm J Cardiol2009103680681119268736
- ShuldinerARO’ConnellJRBlidenKPAssociation of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapyJAMA2009302884985719706858
- SibbingDStegherrJLatzWCytochrome P450 2C19 loss- of-function polymorphism and stent thrombosis following percutaneous coronary interventionEur Heart J200930891692219193675
- SimonTVerstuyftCMary-KrauseMGenetic determinants of response to clopidogrel and cardiovascular eventsN Engl J Med2009360436337519106083
- HarmszeAMvan WerkumJWTen BergJMCYP2C19*2 and CYP2C9*3 alleles are associated with stent thrombosis: a case-control studyEur Heart J201031243046305320833683
- MalekLAPrzyluskiJSpiewakMCytochrome P450 2C19 polymorphism, suboptimal reperfusion and all-cause mortality in patients with acute myocardial infarctionCardiology20101172818720924183
- MegaJLSimonTColletJPReduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysisJAMA2010304261821183020978260
- ParéGMehtaSRYusufSEffects of CYP2C19 genotype on outcomes of clopidogrel treatmentN Engl J Med2010363181704171420979470
- ConnollySJPogueJHartRGEffect of clopidogrel added to aspirin in patients with atrial fibrillationN Engl J Med20093602066207819336502
- US Food and Drug AdministrationFDA announces new boxed warning on Plavix: alerts patients, health care professionals to potential for reduced effectiveness [press release]Silver Spring, MDUS Food and Drug Administration 2010 [March 12]. Available from: http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm204253.htm. Accessed July 27, 2011
- ScottSASangkuhlKGardnerEEClinical Pharmacogenetics Implementation Consortium Guidelines for Cytochrome P450–452C19 (CYP2C19) Genotype and Clopidogrel TherapyClin Pharmacol Ther201190232833221716271
- MegaJLCloseSLWiviottSDCytochrome P450 genetic polymorphisms and the response to prasugrel: relationship to pharmacokinetic, pharmacodynamic, and clinical outcomesCirculation2009119192553256019414633
- KellyRPCloseSLFaridNAPharmacokinetics and Pharmacodynamics Following Maintenance Doses of Prasugrel and Clopidogrel in Chinese Carriers of CYP2C19 VariantsBr J Clin Pharmacol201110.1111/j.1365-2125.2011.04049.x [Epub ahead of print.]
- WallentinLJamesSStoreyRFEffect of CYP2C19 and ABCB1 single nucleotide polymorphisms on outcomes of treatment with ticagrelor versus clopidogrel for acute coronary syndromes: a genetic substudy of the PLATO trialLancet201037697491320132820801498
- KimISJeongYHParkYPlatelet Inhibition by Adjunctive Cilostazol Versus High Maintenance-Dose Clopidogrel in Patients With Acute Myocardial Infarction According to Cytochrome P450 2C19 GenotypeJACC Cardiovasc Interv20114438139121511217
- HwangSJJeongYHKimISCytochrome 2C19 polymorphism and response to adjunctive cilostazol versus high maintenance-dose clopidogrel in patients undergoing percutaneous coronary interventionCirc Cardiovasc Interv20103545045920823393
- ParkKWParkJJLeeSPCilostazol attenuates on-treatment platelet reactivity in patients with CYP2C19 loss of function alleles receiving dual antiplatelet therapy: a genetic substudy of the CILON-T randomised controlled trialHeart201197864164721345843
- PriceMJBergerPBTeirsteinPSStandard- vs high-dose clopidogrel based on platelet function testing after percutaneous coronary intervention: the GRAVITAS randomized trialJAMA2011305111097110521406646
- SaitohTFukushimaYOtsukaHEffects of rabeprazole, lansoprazole and omeprazole on intragastric pH in CYP2C19 extensive metabolizersAliment Pharmacol Ther200216101811181712269976
- ShiraiNFurutaTXiaoFComparison of lansoprazole and famotidine for gastric acid inhibition during the daytime and night-time in different CYP2C19 genotype groupsAliment Pharmacol Ther200216483784611929404
- ShiraiNFurutaTMoriyamaYEffects of CYP2C19 genotypic differences in the metabolism of omeprazole and rabeprazole on intragastric pHAliment Pharmacol Ther200115121929193711736724
- FurutaTOhashiKKamataTEffect of genetic differences in omeprazole metabolism on cure rates for Helicobacter pylori infection and peptic ulcerAnn Intern Med199812912102710309867757
- FurutaTShiraiNTakashimaMEffects of genotypic differences in CYP2C19 status on cure rates for Helicobacter pylori infection by dual therapy with rabeprazole plus amoxicillinPharmacogenetics200111434134811434512
- FurutaTShiraiNTakashimaMEffect of genotypic differences in CYP2C19 on cure rates for Helicobacter pylori infection by triple therapy with a proton pump inhibitor, amoxicillin, and clarithromycinClin Pharmacol Ther200169315816811240980
- TanigawaraYAoyamaNKitaTCYP2C19 genotype-related efficacy of omeprazole for the treatment of infection caused by Helicobacter pyloriClin Pharmacol Ther199966552853410579481
- FurutaTShiraiNWatanabeFEffect of cytochrome P4502C19 genotypic differences on cure rates for gastroesophageal reflux disease by lansoprazoleClin Pharmacol Ther200272445346012386647
- KearnsGLLeederJSGaedigkAImpact of the CYP2C19*17 allele on the pharmacokinetics of omeprazole and pantoprazole in children: evidence for a differential effectDrug Metab Dispos201038689489720223877
- SugimotoMFurutaTShiraiNDifferent dosage regimens of rabeprazole for nocturnal gastric acid inhibition in relation to cytochrome P450 2C19 genotype statusClin Pharmacol Ther200476429030115470328
- SaitohACapparelliEAweekaFCYP2C19 genetic variants affect nelfinavir pharmacokinetics and virologic response in HIV-1-infected children receiving highly active antiretroviral therapyJ Acquir Immune Defic Syndr201054328528919890215
- HaasDWSmeatonLMShaferRWPharmacogenetics of long-term responses to antiretroviral regimens containing Efavirenz and/or Nelfinavir: an Adult Aids Clinical Trials Group StudyJ Infect Dis2005192111931194216267764
- MahgoubAIdleJRDringLGLancasterRSmithRLPolymorphic hydroxylation of Debrisoquine in manLancet19772803858458671400
- EichelbaumMSpannbruckerNSteinckeBDenglerHJDefective N-oxidation of sparteine in man: a new pharmacogenetic defectEur J Clin Pharmacol1979163183187499318
- BradfordLDCYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendantsPharmacogenomics20023222924311972444
- JohanssonILundqvistEBertilssonLDahlMLSjöqvistFIngelman-SundbergMInherited amplification of an active gene in the cytochrome P450 CYP2D locus as a cause of ultrarapid metabolism of debrisoquineProc Natl Acad Sci U S A1993902411825118297903454
- Ingelman-SundbergMGenetic polymorphisms of cytochrome P450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functional diversityPharmacogenomics J20055161315492763
- EckhardtKLiSAmmonSSame incidence of adverse drug events after codeine administration irrespective of the genetically determined differences in morphine formationPain1998761–227339696456
- SindrupSHBrøsenKBjerringPCodeine increases pain thresholds to copper vapor laser stimuli in extensive but not poor metabolizers of sparteineClin Pharmacol Ther19904866866932249379
- StamerUMLehnenKHöthkerFImpact of CYP2D6 genotype on postoperative tramadol analgesiaPain20031051–223123814499440
- WangGZhangHHeFFangXEffect of the CYP2D6*10 C188T polymorphism on postoperative tramadol analgesia in a Chinese populationEur J Clin Pharmacol2006621192793116960721
- KirchheinerJKeulenJTBauerSRootsIBrockmöllerJEffects of the CYP2D6 gene duplication on the pharmacokinetics and pharmacodynamics of tramadolJ Clin Psychopharmacol2008281788318204346
- GascheYDaaliYFathiMCodeine intoxication associated with ultrarapid CYP2D6 metabolismN Engl J Med2004351272827283115625333
- DalénPFrengellCDahlMLSjöqvistFQuick onset of severe abdominal pain after codeine in an ultrarapid metabolizer of debrisoquineTher Drug Monit19971955435449357099
- MadadiPRossCJHaydenMRPharmacogenetics of neonatal opioid toxicity following maternal use of codeine during breastfeeding: a case-control studyClin Pharmacol Ther2009851313518719619
- Kwadijk-de GijselSBijlMJVisserLEVariation in the CYP2D6 gene is associated with a lower serum sodium concentration in patients on antidepressantsBr J Clin Pharmacol200968222122519694742
- BijlMJVisserLEHofmanAInfluence of the CYP2D6*4 polymor phism on dose, switching and discontinuation of antidepressantsBr J Clin Pharmacol200865455856418070221
- LeeSYSohnKMRyuJYYoonYRShinJGKimJWSequence-based CYP2D6 genotyping in the Korean populationTher Drug Monit200628338238716778723
- BertilssonLAberg-WistedtAGustafssonLLNordinCExtremely rapid hydroxylation of debrisoquine: a case report with implication for treatment with nortriptyline and other tricyclic antidepressantsTher Drug Monit1985744784804082245
- BertilssonLDahlMLSjöqvistFMolecular basis for rational megaprescribing in ultrarapid hydroxylators of debrisoquineLancet19933418836638093319
- ThakurMGrossmanIMcCroryDCReview of evidence for genetic testing for CYP450 polymorphisms in management of patients with nonpsychotic depression with selective serotonin reuptake inhibitorsGenet Med200791282683518091432
- CharlierCBrolyFLhermitteMPintoEAnsseauMPlomteuxGPolymorphisms in the CYP2D6 gene: association with plasma concentrations of fluoxetine and paroxetineTher Drug Monit200325673874214639062
- FleemanNDundarYDicksonRCytochrome P450 testing for prescribing antipsychotics in adults with schizophrenia: systematic review and meta-analysesPharmacogenomics J201011111420877299
- DahlMLCytochrome p450 phenotyping/genotyping in patients receiving antipsychotics: useful aid to prescribing?Clin Pharmacokinet200241745347012083975
- SuzukiTMiharaKNakamuraAEffects of the CYP2D6*10 allele on the steady-state plasma concentrations of aripiprazole and its active metabolite, dehydroaripiprazole, in Japanese patients with schizophreniaTher Drug Monit2011331212421157400
- LlerenaABereczRde la RubiaADoradoPQTc interval lengthening is related to CYP2D6 hydroxylation capacity and plasma concentration of thioridazine in patientsJ Psychopharmacol200216436136412503836
- RauTWuttkeHMichelsLMImpact of the CYP2D6 genotype on the clinical effects of metoprolol: a prospective longitudinal studyClin Pharmacol Ther200985326927219037197
- TerraSGPaulyDFLeeCRbeta-Adrenergic receptor polymorphisms and responses during titration of metoprolol controlled release/extended release in heart failureClin Pharmacol Ther200577312713715735607
- JohnsonJAZinehIPuckettBJMcGorraySPYarandiHNPaulyDFBeta 1-adrenergic receptor polymorphisms and antihypertensive response to metoprololClin Pharmacol Ther2003741445212844134
- TerraSGHamiltonKKPaulyDFBeta1-adrenergic receptor polymorphisms and left ventricular remodeling changes in response to beta-blocker therapyPharmacogenet Genomics200515422723415864115
- MürdterTESchrothWBacchus-GerybadzeLActivity levels of tamoxifen metabolites at the estrogen receptor and the impact of genetic polymorphisms of phase I and II enzymes on their concentration levels in plasmaClin Pharmacol Ther201189570871721451508
- MadlenskyLNatarajanLTchuSTamoxifen metabolite concentrations, CYP2D6 genotype, and breast cancer outcomesClin Pharmacol Ther201189571872521430657
- SchrothWHamannUFaschingPACYP2D6 polymorphisms as predictors of outcome in breast cancer patients treated with tamoxifen: expanded polymorphism coverage improves risk stratificationClin Cancer Res201016174468447720515869
- GoetzMPKnoxSKSumanVJThe impact of cytochrome P4502D6 metabolism in women receiving adjuvant tamoxifenBreast Cancer Res Treat2007101111312117115111
- SchrothWAntoniadouLFritzPBreast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypesJ Clin Oncol200725335187519318024866
- LimHSJu LeeHSeok LeeKSook LeeEJangIJRoJClinical implications of CYP2D6 genotypes predictive of tamoxifen pharmacokinetics in metastatic breast cancerJ Clin Oncol200725253837384517761971
- NowellSAAhnJRaeJMAssociation of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patientsBreast Cancer Res Treat200591324925815952058
- WegmanPVainikkaLStålOGenotype of metabolic enzymes and the benefit of tamoxifen in postmenopausal breast cancer patientsBreast Cancer Res200573R284R29015987423
- LashTLLienEASørensenHTHamilton-DutoitSGenotype-guided tamoxifen therapy: time to pause for reflection?Lancet Oncol200910882583319647203
- YuKDHuangAJShaoZMTailoring adjuvant endocrine therapy for postmenopausal breast cancer: a CYP2D6 multiple-genotype-based modeling analysis and validationPLoS One2010512e1564921187922
- HughesSVanderbilt now also routinely gene testing for clopidogrel metabolizer statusTheHeart.Org[website on the Internet]MontrealTheHeart.Org2010 Available from: http://www.theheart.org/article/1139495.do. Accessed August 11, 2011
- RellingMVAltmanRBGoetzMPEvansWEClinical implementation of pharmacogenomics: overcoming genetic exceptionalismLancet Oncol201011650750920413348
- SimSCCYP2D6 allele nomenclature. Home page of the Human Cytochrome P450 (CYP) Allele Nomenclature Committee [website on the Internet] Human Cytochrome P450 (CYP) Allele Nomenclature Committee; 2011 [updated September 16]. Available from: http://www.cypalleles.ki.se/cyp2d6.htm. Accessed September 29, 2011
- CaiWMNikoloffDMPanRMCYP2D6 genetic variation in healthy adults and psychiatric African-American subjects: implications for clinical practice and genetic testingPharmacogenomics J2006634335016550211
- SachseCBrockmollerJBauerSRootsICytochrome P450 2D6 variants in a Caucasian population: allele frequencies and phenotypic consequencesAm J Hum Genet1997602842959012401