 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
The antineoplastic drug bleomycin leads to the side effect of pulmonary fibrosis in both humans and mice. We challenged genetically diverse inbred lines of mice from the Collaborative Cross with bleomycin to determine the heritability of this phenotype. Sibling pairs of mice from 40 lines were treated with bleomycin. Lung disease was assessed by scoring lung pathology and by measuring soluble collagen levels in lavage fluid. Serum micro ribonucleic acids (miRNAs) were also measured. Inbred sibling pairs of animals demonstrated high coinheritance of the phenotypes of disease susceptibility or disease resistance. The plasma levels of one miRNA were clearly correlated in sibling mice. The results showed that, as in humans, the lines that comprise the Collaborative Cross exhibited wide genetic variation in response to this drug. This finding suggests that the genetically diverse Collaborative Cross animals may reveal drug effects that might be missed if a study were based on a conventional mouse strain.
Introduction
The antineoplastic drug bleomycin is now a second-line therapy for certain solid tumors, in part because of its pulmonary toxicity in 10%–20% of patients.Citation1 In about 3% of treated patients, the pulmonary fibrosis that develops is severe and progressive enough that it can lead to death.Citation2–Citation4 It is therefore critical to predict the likelihood of adverse responses to this drug. Bleomycin also causes pulmonary fibrosis in mice and classic inbred strains of mice have been identified that are particularly susceptible (eg, C57BL/6 J, 129, and DBA/2) or relatively resistant (eg, A/J, BALB/c, and C3Hf/Kam).Citation5–Citation7 Indeed, two research groups have begun to map susceptibility loci for bleomycin-induced pulmonary fibrosis in the offspring from crosses between susceptible and resistant lines.Citation7–Citation9 We were interested in a complementary approach of coupling molecular assays to the genetic variation in the Collaborative Cross animals as a way of anticipating adverse drug responses.
While distinct inbred strains of mice or recombinant, congenic strains have been useful in the past for uncovering susceptibility loci for complex traits, we were interested in pursuing a more comprehensive systematic approach, and one that might recapitulate to some extent the levels of human diversity. We thought that our approach might not only lead to the discovery of genetic loci linked to a particular trait, but might also generate candidate biomarkers as part of the process. Here we report the first step towards this goal. We challenged 40 lines from the emerging Collaborative Cross with bleomycin to test the heritability of the phenotype of lung damage and to look for potential biomarkers at the same time. The Collaborative Cross is a panel of recombinant inbred lines of mice derived from eight genetically diverse inbred mouse strains:Citation10,Citation11 A/J, C57BL/6J, 129S1/SvImJ, NOD/LtJ, NZO/HlLtJ, CAST/EiJ, PWK/PhJ, and WSB/EiJ. While each of these lines has been highly inbred, as a group they encompass nearly 90% of the known variation present in laboratory mice.Citation12 The animals used in this study were from the F7–F10 generation of the Oak Ridge National Laboratory stocks, after the eight founding lines were crossbred with one another.Citation11 The Collaborative Cross lines were originally developed for the analysis of complex traits or diseases that arise with contributions from many genes. Since some of the founder strains were known to vary in their sensitivity to bleomycin, we tested whether susceptibility to bleomycin-induced lung damage would be coinherited among sibling pairs that were derived from the founder strains. We also examined the pattern of micro ribonucleic acids (miRNA) in the plasma of affected or resistant animals after treatment. In this model experiment, the genetic diversity of the Collaborative Cross could represent the diversity of the outbred human population while bleomycin stands in for an investigational new drug.
Lung pathology was assessed by biochemical assays and histological scoring, and miRNA were measured in the plasma. Sibling pairs of mice derived from any given Collaborative Cross line showed a high likelihood of coinheritance of the phenotypes related to susceptibility or resistance to lung fibrosis, and some Collaborative Cross lines exhibited extremes of resistance or susceptibility. Plasma miRNA profiles that could be potentially useful as biomarkers of sensitivity or resistance were identified as well. Overall, the results suggested that Collaborative Cross mice are useful for detection of drug-induced side effects, with the advantages that the genetic diversity is potentially higher and that phenotypes may be more extreme. Because of large genetic diversity encompassed within the Collaborative Cross or their derived outbred population, the Diversity Outcross (Pers comm; E Chesler, 2011) represents a powerful way to anticipate the possible side effects of investigational new drugs.
Materials and methods
Generation of recombinant inbred lines of mice
The Collaborative Cross combines the genomes of eight genetically diverse founder strains:A/J, C57BL/6 J, 129S1/SvImJ, NOD/LtJ, NZO/HlLtJ, CAST/EiJ, PWK/PhJ, and WSB/EiJ. These strains of mice were systematically and independently outcrossed to bring together all eight genomes, and then inbred.Citation10,Citation11 The Collaborative Cross progenitor lines included founder lines previously shown to be susceptible to bleomycin-induced pulmonary fibrosis (129, C57BL/6) as well as a founder line shown to be relatively resistant (A/J).Citation1,Citation5,Citation6,Citation9 A randomly chosen subset of 40 lines was used in this study.
Dosing with bleomycin
Two male mice 6–8 weeks of age were selected from each of 40 inbred lines of the Collaborative Cross. Animals were dosed intraperitoneally with 0.035 units of bleomycin/g body weight on each of 8 days spread over the next 33 days.Citation13 Animals were dosed on days 1, 4, 8, 11, 15, 18, 22, 25. On day 33, blood plasma was collected for miRNA analysis. Mice were subsequently euthanized and the right lungs were placed in 0.5 M acetic acid (for assay of soluble collagen) and the left lungs were gently inflated and placed in 10% buffered formalin for histology. Tail DNA was prepared for high-density genotyping from all animals.
Semiquantitative scoring of lung fibrosis
Fixed lungs were prepared for histologic examination and semiquantitative assessment of inflammation and fibrosis. One section from one lung of each mouse was stained with hematoxylin and eosin while the other was stained with Masson’s trichrome for visualization of collagen. Each section was scored by a board-certified pathologist for inflammation, macrophage accumulation in alveoli, and fibrosis using a semiquantitative scale of 0 (no pathologic changes) to 4 (marked pathology approaching maximal). Semiquantitative assessment of each lesion was based on the presence of rare individual affected foci (minimal, grade 1), increased frequency and size of individual lesions with occasional confluence (mild, grade 2), large areas of involvement and confluence (moderate, grade 3), or complete lung involvement with the lesion in question (marked, approaching maximal, grade 4). Internal peer review was performed by another board-certified pathologist to confirm findings and obtain consensus as to interpretations.
Soluble collagen assay
Soluble collagen was measured by dye binding to gly-X-Y collagen fragments (Sircol kit; Accurate Chemical Corp, Westbury, NY). Collagen concentrations were inferred by absorbance at 540 nm following the manufacturer’s instructions. Data were reported as mg collagen per 25 mL volume analyzed.
Statistical analysis of data
Members of each sibling pair were randomly assigned to one of two groups such that the two members were in different groups. If a third sample was obtained from a given line, one of the three samples was randomly omitted. To test for genetic concordance of pathology scoring, the odds ratio of concordant vs discordant pairs was obtained.
A 95% confidence interval was obtained for each natural logarithm of the odds ratio, and P values to test whether this value differed from zero (ie, that the observed responses were concordant among sibling pairs) were obtained. Results are presented in .
Table 1 Soluble collagen data and semiquantitative pathology scores
A sibling pair regression analysis (performed using SYSTAT[v 12; Systat Software, Inc, Chicago, IL]) is an estimate of genetic correlation, and a significant correlation implies that a common source of genetic variance among the population is responsible for the pairs’ phenotypic value. In a conventional sibling pair regression, individuals are assumed to be 25% similar and therefore, the regression coefficient is multiplied by four to extrapolate to heritability in a randomly mating wild population. Because the siblings are from somewhat inbred stocks, this would yield an upwardly biased estimate and because we are not attempting to estimate the true population heritability for this biomedically relevant phenotype, we used the conservative raw coefficient.
Plasma miRNA profile
miRNA levels were measured in heparinase-treated blood plasma as described by a slight modification of a previously published reverse transcription polymerase chain reaction protocol.Citation14 For studies of individual miRNAs, total RNA was extracted from previously frozen mouse plasma using Trizol reagent (Qiagen, Germantown, MD) and purified using a miRNEASY kit (Qiagen) according to the manufacturer’s instructions. The expression level of mature miRNAs was determined by real time polymerase chain reaction analysis following stem-loop reverse transcription with Taqman miRNA assay reagents (Applied Biosystems, Carlsbad, CA). The amplification program included an initial activation and denaturation at 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds, 60°C for 1 minute. The relative amounts of miRNAs were normalized to an equal input volume of plasma. After the reaction, cycle threshold values were calculated using fixed manual threshold settings. A larger group of miRNAs was profiled to test for an association with pathology scores. In this case, previously frozen blood plasma samples were treated by adding a one-tenth volume of 1 mg/mL Heparinase I (in 20 mMTris-HCl, pH 7.5, 50 mMNaCl, 4 mM CaCl2, and 0.01% bovine serum albumin; Sigma-Aldrich, St Louis, MO) and incubated for 1 hour at room temperature before reverse transcription. Megaplex primers for Taqman miRNA profiling (Applied Biosystems) were used to preamplify samples prior to loading the samples into Taqman low density array miRNA rodent card A, version 2 (Applied Biosystems). The reverse transcriptase polymerase chain reaction was carried out according to manufacturer’s instructions. The amplification program included initial activation and denaturation at 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds, 60°C for 1 minute. Levels for each miRNA were normalized to the levels of U6 small nuclear RNA which was measured in parallel.
Results
In this study two male sibling mice from each of 40 Collaborative Cross lines were treated with eight doses of Bleomycin over a period of 1 month. At the end of the dosing period all mice were euthanized, blood samples were collected to prepare plasma, lungs were dissected for an investigation of the lung pathology, and tail snips were saved to isolate DNA for genotyping.
The larger lobe from each lung was processed for histologic examination and soluble collagen was assayed in the smaller lobe. The histologic examination of stained lung sections from each Collaborative Cross animal showed that some animals showed no obvious lung damage while others displayed extreme fibrotic lung disease, consistent with the phenotypes of both resistant and susceptible lines in the breeding scheme. As shown in , the lungs of some animals appeared normal, and we noted varying levels of pathology among others. Each lung was scored semi-quantitatively for inflammation, fibrosis, and accumulation of macrophages, and then also given an overall score for bleomycin-induced pathology encompassing the aforementioned changes. Normal lungs were assigned a score of 0 based on the absence of these pathologic signs. Grade 1 lungs showed occasional foci with active inflammation including infiltrating macrophages and a few sites with excess collagen deposition. Lungs scored as grade 2 showed more confluent areas of inflammation with fibrosis and more frequent foci of macrophage accumulation. Lungs scored as grade 3 showed large areas of inflammation, sometimes including the entire lung, with extensive fibrosis and in some cases the emergence of cells with the appearance of myofibroblasts. Deposition of collagen in the fibrotic regions was confirmed in grade 2 and 3 lungs after serial sections were stained with Masson’s trichrome (data not shown). The semiquantitative scores for lung pathology are summarized in .
Figure 1 Representative lung photomicrographs. Typical images that were used for semiquantitative assessment of lung pathology are shown (4 × magnification). Upper left: normal, mouse/slide 81, MouseTrack #193319; Upper right: grade 1, Mouse/slide 34, MouseTrack #184942; Lower right: grade 2, mouse/slide 74, MouseTrack #192908; Lower left: grade 3, mouseslide 73, MouseTrack #192907.
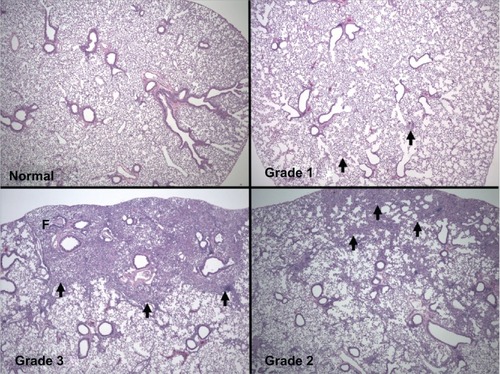
The lines of mice in our study were inbred for four to seven generations by following a defined funnel inbreeding scheme. From this degree of inbreeding, 65%–80% of genome segments in sibling pairs were predicted to be identical by descent.Citation11 We thus asked to what extent sibling animals showed a tendency to coinherit the presence or absence of any lung pathology. Since the pathology scores were obtained before we knew the identity of the sibling pairs, we first asked if mice with similar phenotypes were siblings. A preliminary analysis of the scores in showed that mice with mild lung pathology were more likely to have siblings with mild pathology than they were to have discordant siblings (P 0.86 > P 0.79), which is suggestive of a genetic contribution to the disease. This relationship was noted especially for inflammation and fibrosis in the lung. Likewise, mice unaffected by the drug treatment were also more likely to have siblings that were unaffected than affected siblings, which is again suggestive of a genetic contribution to disease resistance. The details of the probability analysis are presented in and .
Table 2 Pathology features and numbers of cases observed
Table 3 Probabilities of different observed pathologies
To extend these findings, we next calculated the relative risk that if a given animal was affected, that the sibling would be affected. We did this for both affected animals (scores > 0) and severely affected animals (scores > 1). For example we noted that mildly affected probands were 2.45 to 2.9 times more likely to have an affected sibling than unaffected probands on measures of macrophages, inflammation, and fibrosis. The relative risk of severe inflammation or excessive macrophages among siblings of affected probands was 1.8 and 1.7 times that of unaffected probands. The most striking finding was that the relative risk for severe fibrosis given an affected sibling was 12.75 times that for those with unaffected siblings. Our analysis also showed a significant concordance of inflammation (odds ratio [OR]: 7.13;P < 0.01) or a macrophage response (OR: 12.00; P < 0.01) in mildly affected animals with affected siblings. In severely affected animals with affected siblings there was a significant concordance of severe fibrosis (OR: 23.25, P < 0.01) as well as inflammation (OR: 7.14, P < 0.04). These results are presented in more detail in and . Thus, mice with either no response or a moderate or severe response to bleomycin treatment tended to have siblings that shared these responses, which is readily explained if there is a strong genetic component to the phenotype of resistance or susceptibility.
Table 4 Relative risk calculations for affected compared to unaffected animals
Table 5 Relative risk calculates for severely affected compared to unaffected animals
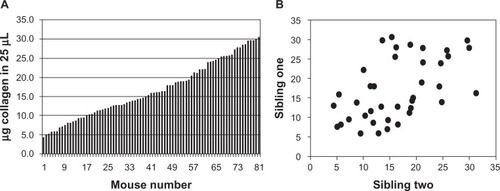
Collagen metabolism is active in the fibrotic lung because of the degradation of normal lung structures as well as the synthesis of fibrillar collagens as a component of new extracellular matrix. We thus measured soluble collagen in the lungs of all animals as another independent test of a potentially heritable aspect of the response to bleomycin (). When simply ranked numerically, soluble collagen scores varied over a six-fold range (, panel A) and when the value for one sibling was regressed on the value of its sibling, strong genetic correlation was observed with a regression coefficient of 0.544 (, panel B). Sibling mice with similar soluble collagen scores were noted throughout the range of observed soluble collagen values and overall disease susceptibility to resistance, consistent with a genetic basis for these responses that segregated into defined inbred lines of mice.
Figure 2 Inheritance analysis of soluble collagen levels in the lungs. Panel A: lung soluble collagen values from all animals. Panel B: regression analysis of soluble collagen levels for 39 sibling pairs of mice. The data show the tendency for the concentration of collagen that was extracted from the lungs to be coinherited, with a regression coefficient of 0.544.
We also profiled selected miRNA species in blood plasma samples, looking first at miRNA that had been reported to have roles in lung disease. MiR-34a was readily detected, as shown in . When the levels of this miRNA were plotted from one mouse as a function of the level in its sibling (), a regression coefficient of 0.58 was obtained, suggesting that the plasma levels showed evidence of genetic correlation. In contrast, a second miRNA, miR-21, showed similar plasma levels, but the levels in siblings showed little correlation ( and ). Four more miRNAs that are known to be expressed in the lung showed little correlation between sibling pairs of mice, at least when they were measured in the plasma. The correlation coefficients of these miRNAs are presented in . Some miRNAs were elevated or decreased in the plasma of animals with the most severe lung pathology, but these data did not quite approach the level of statistical significance (not shown).
Figure 3 miRNA levels in plasma. A: the plasma levels of miR-34a measured in 70 mice; B: data from A replotted by sibling pairs; C: the plasma levels of miR-21 measured in 67 mice; D: data from C replotted by sibling pairs.
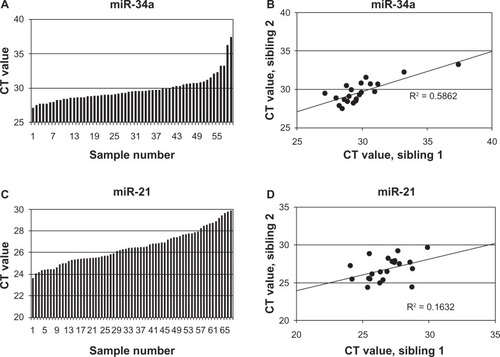
Table 6 Regression coefficients for coinheritance by siblings of miRNA peripheral blood levels
Discussion
In this study we found that in response to a drug challenge a group of genetically diverse mice exhibited the same side effects as humans. Since some of these different lines of the Collaborative CrossCitation10,Citation11 were resistant to the drug challenge, while others were extremely sensitive, our study recapitulated the variability in human responses that have been observed with this drug. A study based on other recombinant inbred lines such as the BXD which are derived from strains (C57BL/6; DBA/2) that are both quite susceptible to bleomycin-induced pulmonary fibrosis would not have been as informative. The new highly diverse genetic reference and mapping populations made from the eight strains of Collaborative Cross are useful resources to dissect this phenotypic variation.
Our data showed that the phenotype of fibrotic lung disease in response to the drug bleomycin was more likely in a given animal if a sibling was also affected. An independent measure of lung fibrosis, soluble collagen levels derived from the lung, showed the same effect. In our semiquantitative scoring, not all histological features we looked at showed clear evidence of heritability (ie, the presence of inflammation, ) but we did not stain for other features that were likely to have been quite heritable, such as myofibroblastsCitation15 (see for example , grade 3 disease). Another source of error is the fact that we only examined two 5 μm sections from a lung that is 5 mm thick. Examining and averaging multiple sections per lung could have given more accurate scores, and more highly quantitative metrics may have led to better detection of covariation. Nonetheless the coinheritance pattern that emerged for the phenotype of destructive lung disease is most easily explained as having a genetic basis that segregated among the 40 recombinant inbred mouse lines we studied.
We also profiled miRNA in blood plasma. We found the plasma levels of at least one miRNA varied among all strains tested, but the level in one sibling was predictive of the level in the other sibling, as might be expected if the expression level for this miRNA (miR-34a) was segregating as a heritable trait. But the levels of several other miRNAs were not closely matched in sibling mice. We were also unable to find a statistically reportable association between the plasma miRNA profile and lung disease. Recent work shows that the lung and the blood profile of miRNAs change when single inbred mouse strains are treated with bleomycin (Wang K, Cho JH, Marsh C, et al; unpublished observations) or in mice with destructive lung disease that are deficient for adenosine deaminase (Blackburn M, Cho JH, Wang K, et al; unpublished observations). We suspect that technical problems arising from small sample size and the presence of heparin in the plasma samples hampered the miRNA quantitation in this study. MiRNA profiles in the blood changed dramatically, when mice were challenged with intraperitoneal acetaminophen.Citation14 Despite this, blood miRNA profiles may still be worth pursuing in drug toxicity studies since the changing profile in the blood may have biomarker information and because the particular miRNAs detected may participate directly in the disease process.Citation15,Citation16
In this study, about 8 months were required to generate the mice while the drug exposure and data analysis added 2–3 months. When the full spectrum of the final Collaborative Cross lines of mice, inbred to a satisfactory degree, are all available at the same time from one source, such a study could be completed in a fraction of this time, perhaps 3–4 months or less. This is about the same time required for a standard series of preclinical rodent toxicity tests, which are applied to all new novel chemical entities or investigational drugs. Studying mice from the highly diverse Collaborative Cross at the same time or in addition to conventional dose escalation studies in rodents should reveal rare phenotypes or reactions that might be genetically determined and thus entirely missed if only one or two classic laboratory mouse strains were used. Alternatively, this can be done now in the Diversity Outcross recombinant inbred lines available from The Jackson Laboratory (Pers comm; Chesler E, 2011).
The genetic diversity of the Collaborative Cross has been estimated to rival that of the outbred human population.Citation12 If a drug phenotype is heritable in selected inbred lines of the Collaborative Cross as we demonstrated for the destructive lung disease secondary to bleomycin ( and ), the inbred lines thus identified can be used to uncover the genetic basis of the response in follow-on studies. Since all of the inbred lines of the Collaborative Cross will have genotypes and even imputed sequence information publicly available, the identification of a pattern of markers that define a region of the genome that segregates with the phenotype should be simplified as would the identification of candidate genes or loci that contribute to the phenotype. Use of genetically diverse mice offers a new way to understand the biological basis of expected or unexpected drug effects in great detail.
For some time, variations in the distribution or structure of the intracellular peptidase called bleomycin hydrolase that can degrade the drug were thought to underlie the induced fibrosis in mice and men.Citation1 More recently, in addition to lung fibrosis, it is now recognized that subcutaneously injected bleomycin can cause a scleroderma-like condition with autoimmune attributes.Citation17,Citation18 This model of human systemic scleroderma suggests that an early step in the catabolism of bleomycin results in the generation of reactive oxygen species in vascular endothelial cells and smooth muscle causing these cells to die by apoptosis or necrosis.Citation19 Dead or dying cells liberate hyaluronan and HMGB-1 protein that can activate B-cells, CD4 T cells, and possibly macrophages via toll receptors TLR2 and TLR4 to produce profibrogenic cytokines such as IL-6 and TGF-β, resulting in local fibrosis. B cells may infiltrate and persist in lung or skin via CD19-dependent signaling.Citation18,Citation20 Transfer of CD4 cells from an inflamed animal will transfer scleroderma to a naïve animal.Citation21 The genetic bases of all aspects of this proposed cascade are amenable to confirmation and exploration by using the approach outlined in this study.
We have demonstrated that when the mouse lines of the Collaborative Cross were challenged with a drug, a pattern of adverse events was evoked that closely modeled disease susceptibility in humans.Citation2–Citation4 We suggest that by avoiding the use of single inbred lines of mice and instead working with a group of lines that were deliberately bred to encompass an extremely high level of genetic diversity, low probability but high consequence drug effects can be revealed in a single experiment. Moreover, for any given phenotype that might arise in such a study, the path to discover the genetic basis for the phenotype is facilitated. Increasingly, there is little excuse for not knowing the full spectrum of adverse events that may be associated with an investigational new drug that can be revealed in a mouse, long before the start of human trials.
Acknowledgements
The authors thank Christine Bhirdo, Lisa Branstetter, Pat Hunsicker, William Parkhurst, Jason Spence, Ginger Shaw, and Sarah Shinpock at the Oak Ridge National Laboratory for the production, treatment, and dissection of the Collaborative Cross mice. We also thank our colleague at Battelle: Connie Essman-Wood, at the Ohio State University: Melissa Piper, and colleagues at the Institute for Systems Biology: David Baxter and Ji-Hoon Cho. The authors wish to acknowledge support from the Pulmonary Systems Biology Initiative from the Battelle Memorial Institute (to Oak Ridge National Laboratory and Institute for Systems Biology), from the University of Luxembourg and the Luxembourg Centre for Systems Biomedicine (to the Institute for Systems Biology), and the Systems Biology Center Grant (NIH-GM 076547) to the Institute for Systems Biology. The Collaborative Cross was initiated with the generous support of The Ellison Medical Foundation.
Disclosure
The authors report no conflicts of interest in this work.
References
- LazoJSHoytDGThe molecular basis of interstitial pulmonary fibrosis caused by antineoplastic agentsCancer Treat Rev1990172–31651671703044
- LeviJRaghavenDHarveyVThe importance of bleomycin in combination chemotherapy for good-prognosis germ cell carcinomaJ Clin Oncol1993117130013057686216
- SimpsonABPaulJGrahamJKayeSBFatal bleomycin pulmonary toxicity in the west of Scotland 1991–1995: a review of patients with germ cell tumorsBr J Cancer1998788106110669792151
- SleijferSBleomycin-induced pneumonitisChest2001120261762411502668
- SchrierDJJunkelRGPhanSHThe role of strain variation in murine bleomycin-induced pulmonary fibrosisAm Rev Respir Dis1983127163666185026
- EkimotoHTakadaKOhnukiTDifferent sensitivity to bleomycin-induced pulmonary fibrosis among various strains of miceJ Clin Biochem Nutr1987212531
- HastonCKWangMDejournettREBleomycin hydrolase and a genetic locus within the MHC affect risk for pulmonary fibrosis in miceHum Mol Genet200211161855186312140188
- BarthRKHanchettLABaecher-AllanCMMapping susceptibility genes for the induction of pulmonary fibrosis in miceChest2002121Suppl 321S11893659
- LemayAMHastonCKBleomycin-induced pulmonary fibrosis susceptibility genes in AcB/BcA recombinant congenic micePhysiol Genomics2005231546116179420
- ChurchillGAAireyDCAllayeeHThe Collaborative Cross, a community resource for the genetic analysis of complex traitsNat Genet200436111133113715514660
- CheslerEJMillerDRBranstetterLRThe Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems geneticsMamm Genome200819638238918716833
- RobertsAPardo-Manuel de VillenaFWangWMcMillanLThreadgillDWThe polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems geneticsMamm Genome2007186–747348117674098
- BaranCPOpalekJMMcMakenSImportant roles for macrophage colony-stimulating factor, CC chemokine ligand 2, and mononuclear phagocytes in the pathogenesis of pulmonary fibrosisAm J Respir Crit Care Med20071761788917431224
- WangKZhangSMarzolfBCirculating microRNAs, potential biomarkers for drug-induced liver injuryProc Natl Acad Sci U S A2009106114402440719246379
- ChoJHGelinasRWangKSystems biology of interstitial lung diseases: integration of mRNA and microRNA expression changesBMC Medical Genomics20114821241464
- PanditKVCorcoranDYousefHInhibition and role of let-7d in idiopathic pulmonary fibrosisAm J Respir Crit Care Med2010182222022920395557
- D’CruzDAutoimmune diseases associated with drugs, chemicals and environmental factorsToxicol Lett2000112–113421432
- YoshizakiAIwataYKomuraKCD19 regulates skin and lung fibrosis via toll-like receptor signaling in a model of bleomycin-induced sclerodermaAm J Pathol200817261650166318467694
- IshikawaHItoSNishioNYuzawaYMatsuoSIsobeKInjection of bleomycin in newborn mice induces autoimmune sialitis that is transferred by CD4 T cellsImmunol Cell Biol200987435135819204734
- KomuraKYanabaKHorikawaMCD19 regulates the development of bleomycin-induced pulmonary fibrosis in a mouse modelArthritis Rheum200858113574358418975313
- IshikawaHTakedaKOkamotoAMatsuoSIsobeKInduction of autoimmunity in a bleomycin-induced murine model of experimental systemic sclerosis: an important role for CD4+ T cellsJ Invest Dermatol200912971688169519158840