Abstract
Background:
A number of research studies on the genetics of opiate dependence have focused on the μ-opioid receptor (OPRM1), which is a primary target for opiates. This study aims to identify genetic polymorphisms within the OPRM1 gene involved in response to the biopsychosocial treatment in opiate-dependent individuals of Arab descent.
Methods:
Unrelated Jordanian Nationals of Arab descent (N = 183) with opiate dependence were selected for this study. These individuals, all males, met the DSM-IV criteria for opiate dependence and were undergoing a voluntary 8-week treatment program at a Jordanian Drug Rehabilitation Centre. All individuals were genotyped for 22 single nucleotide polymorphisms (SNPs) within the OPRM1 gene using the Sequenom MassARRAY® system (iPLEX GOLD). Statistical analyses were carried out using the R package.
Results:
Patients receiving biopsychosocial treatment showed that there was a significant difference in their OPRM1 SNPs’ genotyping distribution between good, moderate, and poor responders to the treatment at two sites (rs6912029 [G-172T], and rs12205732 [G-1510A], P < 0.05, Fisher’s exact test).
Conclusion:
This study is the first report of an association between the OPRM1 G-172T and G-1510A polymorphisms and treatment response for opiate dependence. Specifically, this study demonstrated that the OPRM1 GG-172 and GG-1510 genotypes were more frequent among patients who were nonresponders to the biopsychosocial treatment. However, further pharmacogenetic studies in a larger cohort of opiate-dependent patients of Arab descent are needed to confirm these findings and identify individuals with increased chance of relapse.
Introduction
Opiates are considered to be among the most addictive illicit drugs.Citation1 Anthony et alCitation2 reported that 23% of individuals who use opiates at least once in their lifetime become dependent, compared to17% for cocaine and 13% for other illicit drugs. Opiate dependence has the highest propensity for causing physical harm to the user, and societal harm through damage to family and social circles. The economic costs of opiate dependence are also high and include the costs of health care, social care, and crime.Citation3–Citation6
Currently, the three major maintenance pharmacotherapies for the treatment of opioid dependence are methadone, buprenorphine, and naltrexone.Citation7 When combined with psychosocial services, these maintenance treatments are effective in reducing opiate use, dangerous behavior, and criminal activity, while improving the mental health of patients.Citation6,Citation7 However, the majority of opiate-dependent individuals remain out of treatment, and those who remain in treatment exhibit only 60%–70% success rates.Citation6 Therefore, it remains an essential goal to further the understanding of the factors underlying poor treatment outcomes and assist in the development of more individualized, optimized, and ultimately more effective treatments for opiate drug dependence.
Pharmacogenetic studies have shown that both methadone and buprenorphine display a large interindividual variability in their pharmacokinetics and contribute to high interindividual variability in response to opiate-dependence treatment.Citation7–Citation9 Meta-analyses of many studies of naltrexone hydrochloride have also suggested that the effect size for response over placebo is in the small-to-moderate range.Citation10–Citation14 Several genetic studies have suggested that naltrexone does not have therapeutic effects in some alcohol-dependent individuals.Citation14–Citation17 Human laboratory studies have also reported that alcohol increases endogenous opioids usage more in patients with a family history of alcohol dependence.Citation18 Apart from alcohol dependence, little attention has been devoted to the possible genetic factors affecting the treatment response to naltrexone in opiate drug dependence. However, demonstrations of the role of the brain’s opioidergic system in mediating drug tolerance and dependence have identified it as a potential source of interindividual variability in the pharmacodynamics’ response to opioids (eg, heroin, buprenorphine, and methadone) and opiate antagonists such as naltrexone.Citation14–Citation17
Research into the genetics of opiate dependence has focused on the opioidergic system, which is the primary target for opiates – in particular, heroin.Citation7,Citation19 Heroin is converted into morphine in vivo and activates the opioid receptors (μ, δ, κ).Citation7,Citation20,Citation21 This modulates several physiological processes such as pain, reward, nociception, immune, and gastric function, and stress and treatment responses.Citation7,Citation20,Citation21 The opioid receptor μ 1 (OPRM1) is thought to account for most of the opioidergic effects.Citation20–Citation22 OPRM1 is also the primary site of action of endogenous ligands such as β-endorphin and enkephalinCitation23 and μ-opioid receptor antagonists such as naltrexone,Citation24 agonists such as methadone,Citation25 or partial antagonists such as buprenorphine.Citation20 Haile et alCitation19 reported that opiates’ physiological and subjective effects are associated with enhanced release of β-endorphin.
The OPRM1 gene (6q24-q25; Gene ID: 4988) encodes the μ opioid receptor, which is widely distributed in the brain.Citation26–Citation28 The OPRM1 receptor is a membrane of the G protein-coupled receptor family,Citation23,Citation29 and over 300 OPRM1 sequence variants have been identified to date.Citation30,Citation31 The A118G Single-nucleotide polymorphism (SNP) in exon 1 of the OPRM1 gene is the SNP most frequently found in the human population.Citation32 This polymorphism encodes an Asn40Asp amino acid substitution that appears to be associated with changes in function. Bond et alCitation33 reported that β-endorphin has a higher binding affinity (threefold) at the Asp40 mutated receptor than at the receptor encoded by Asn40.Citation33,Citation34 In addition, β-endorphin activates G protein-coupled inwardly rectifying potassium channels (GIRKs) more in the presence of the Asp40 allele than the Asn40 allele.Citation33,Citation34 However, other studies have reported that the binding affinity or potency of β-endorphin for the variant receptor is not different from the normal receptor.Citation23,Citation33–Citation35 A recent meta-analysis reported that the OPRM1 Asn40Asp does not appear to affect risk for drug dependence,Citation36 but other studies report that it may influence response to opioid antagonist treatment for alcohol dependence using naltrexone.Citation37
Various studies have provided varied and conflicting results for an association between opioid receptor gene polymorphism and treatment response.Citation35–Citation41 For example, Oslin et al found that individuals with the Asp40 allele had significantly lower rates of relapse and took longer to resume heavy drinking than Asn40/Asn40 homozygous in a sample of 130 European American alcoholics receiving naltrexone as treatment.Citation42 A similar result has been found in Korean alcoholics with a higher therapeutic effect of naltrexone in individuals who had the Asp40 variant genotype compared to the Asn40 genotype.Citation39
As very few pharmacogenetics studies have been conducted on the treatment of opiate dependence, such studies are required to better understand how opiate addicts respond to specific drug treatments. This study aimed to identify genetic variations within the OPRM1 gene involved in responsiveness to the biopsychosocial treatment in Jordanian opiate-dependent patients of Arab descent.
Materials and methods
Study patients
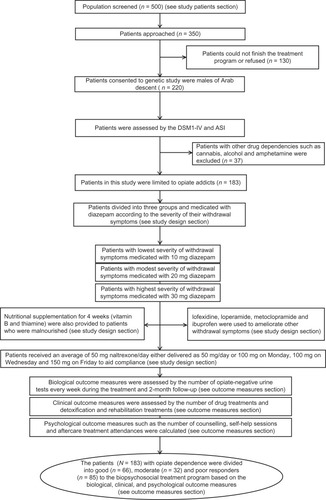
Patients for this study were recruited from inpatient and out-patient programs at the National Centre for Rehabilitation of Addicts (NCRA) at the Jordanian Ministry of Health and the Drug Rehabilitation Centre at the Jordanian Public Security Directorate (DRC-PSD). Inclusion criteria were having a diagnosis of opiate dependence and being unrelated Jordanian Arab males and between 18 and 58 years of age. The Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV) was used to assess the medical and psychiatric status of patients.Citation43 Psychiatric diagnosis was established using a structured baseline interview, which was based on the Addiction Severity Index (ASI) criteria.Citation44 The clinical diagnoses and structured clinical interviews were conducted by independent psychiatric consultants. Patients were excluded if any of the following applied: axis-I comorbidity such as diagnosis of schizophrenia, major depression, bipolar I and II disorder, schizoaffective disorder, a serious medical illness, or those receiving psychotropic medications. Those patients with serious medical conditions such as neuroendocrine, metabolic, or cardiovascular diseases, neurodegenerative disorders, or epilepsy were also excluded. If patients used more than one substance, they were included only if their major drug dependence was an opiate. Initially, 500 patients were screened (see ). Based on the inclusion and exclusion criteria mentioned above, 350 patients were approached to participate in this study. Of these patients, 130 patients could not finish the treatment program for clinical reasons. Of the remaining patients, 220 agreed to be part of this study. A further 37 (16%) patients were then excluded from the final analysis due to the type of drug dependence and the samples were limited to only opiate dependence. In total, complete data were obtained from 183 patients with opiate dependence.
Figure 1 Flow chart of subjects, treatment approach, and outcome measures.
This study was conducted according to the provisions of the Australian Medical Association Code of Ethics and the World Medical Association Declaration of Helsinki. The study was also subject to, and in compliance with, the National Statement on Ethical Conduct in Human Research, Australia (2007). Ethical approval to conduct this research was granted by the Human Research Ethics Committee of The University of Western Australia (Ref No RA/4/1/4344). This study was also approved by the Human Ethics Committee of the Jordanian Ministry of Health (Ref No Development/Trainees/535) and by the Institutional Review Board of the Jordan University of Science and Technology (Ref No RA/16/1/2010). Written informed consent was obtained from all patients before participation in the study.
Study design
At the initial screening, several procedures were undertaken to collect demographic and clinical data. These included capturing demographic data such as date of birth, sex, nationality, marital status, children, and occupation. A medical history including drug overdose and suicide attempts was also taken from the participants’ medical records. Information on substance abuse was also collected from their medical records including type of substance, cause of addiction, initial date of addiction, starting and last taken amount of opiate, route of administration, withdrawal periods, and hospitalizations due to opiate abuse. Data on smoking status and blood relatives with a history of drug abuse were also collected from participarts’ medical records. All data were coded and no specific individual was identified.
Treatment approach
All subjects who met the DSM-IV criteria for opiate dependence received biopsychosocial treatment in the NCRA and DRC-PSD programs (see ). Treatment consisted of 7-day inpatient detoxification using a regime of diazepam medication and 7-week oral naltrexone as maintenance treatment. This is often accompanied by withdrawal symptoms and occasionally fatal side effects.Citation7 Four other oral medications (lofexidine, loperamide, metoclopramide, and ibuprofen) were used to ameliorate other withdrawal symptoms such as stomach cramps and diarrhea, and nutritional supplementations (vitamin B and thiamine) were provided for 4 weeks. The NCRA and DRC-PSD treatment program also provided patients with psychosocial support. Inpatient groups were offered counseling sessions three times a week for a total of 8 weeks. Each session lasted for 2 hours. In addition, patients participated in 1-hour individual counseling sessions per week over the 8-week treatment program. These participants were described as patients completing treatment and continuing with aftercare programs.
In the NCRA and DRC-PSD programs, chlordiazepoxide is prescribed for outpatients as it has a lower abuse potential. Valium (diazepam) is used for inpatients as it has a faster action and a higher dose effect. All patients are given four doses of diazepam every day at early morning, midday, early evening, and at bedtime and reviewed daily to assess withdrawal symptoms. On admission, they are given four separate doses of 30 mg. The next day’s planned dosages are based on ongoing assessment of the patients’ symptoms rather than the length of the course or the prescribed starting dose. The patients recruited for the study were divided into three groups according to the severity of their withdrawal symptoms. The first group of patients (lower-dose responders) had the lowest severity of withdrawal symptoms and were medicated with 10 mg diazepam four times a day. The second group of patients (high-dose responders) had moderately severe withdrawal symptoms and were medicated with equal to or greater than 20 mg diazepam four times a day. The third group of patients (high-dose nonresponders) consisted of nonresponders to treatment with severe withdrawal symptoms and were medicated with equal to or greater than 30 mg diazepam four times a day. Four other oral medications were used to ameliorate other withdrawal symptoms: lofexidine, loperamide, metoclopramide, and ibuprofen. Lofexidine, an α-adrenergic agonist, was used to reduce the other withdrawal symptoms such as chills, sweating, stomach cramps, diarrhea, muscle pain, runny nose, and watering eyes. These patients, underwent, a 10-day course of lofexidine treatment starting at 0.2 mg two to four times a day and increasing in units of 0.4 mg to a maximum of 2.4 mg. Loperamide was used for treatment of diarrhea. All patients were given an initial dose of 4 mg after each loose stool up to a total of 16 mg per day. Metoclopramide was used for the treatment of nausea and vomiting. Patients were prescribed 10 mg/day up to a maximum of 30 mg/day. Ibuprofen was used to reduce fever and headaches and treat muscle pains. Patients were given an initial dose of 0.4 mg of ibuprofen every 4 to 6 hours, which was increased to a maximum daily dose of 1.6 mg according to their response and tolerance to the drug. Supplementary vitamins (eg, vitamin B and thiamine) were also provided to patients who were malnourished.
For the patients who had successfully completed the detoxification and withdrawal period and who had been opiate free for more than 7 days, the opiate antagonist, naltrexone, was used as an aid to prevent relapse and promote abstinence. The drug was given at a starting dose of 25 mg/day, increased to 50 mg/day. Sometimes, patients received an average of 50 mg naltrexone/day either delivered as 50 mg/day or 100 mg on Monday, 100 mg on Wednesday, and 150 mg on Friday to aid compliance.
Patients completing the intensive program are offered an aftercare program, which includes one individual counseling session per week for up to 6 months. The treatment approach is problem-oriented and focuses on achieving well-defined goals such as the availability, accessibility, affordability, and efficiency to all substance abuse patients in need of treatment. These objectives are outlined in a treatment plan that is prepared at admission and is updated for each new treatment program. Medical, behavioral, supportive, and relapse prevention strategies are drawn from different treatment models.Citation7 In this study, the treatment team was kept blind to the genetic status of the patients.
Outcomes measures
A semi-structured baseline interview was developed from the ASI criteria. These interviews were used to collect the demographic and clinical data of the 183 patients. The family history of substance abuse was also obtained. This allowed the subset of drug-dependent patients whose addiction may be influenced by genetic factors to be identified.Citation45 Berrettini and Persico suggest that the likelihood of detecting susceptibility genes is higher in these individuals.Citation46 In-treatment, end-of-treatment, and follow-up assessments were also conducted to provide valid estimates of opiate abstinence. Retention and attrition from treatment were also recorded (see ).
The measures to assess outcome were the number of negative urines. Urine drug screening (UDS) was performed for all patients at the NCRA and DRC-PSD on the day of admission to the treatment program. Every week during the treatment, after treatment completion and each follow up, the patients were randomly screened for drugs using The Multi Drug Test 10 Panel (Jant Pharmacal Corporation, Encino, CA). This test is a one-step immunoassay for the detection of cannabis, cocaine, phencyclidine, opiates, methamphetamine, methadone, amphetamine, barbiturates, benzodiazepines, and oxycodeine. The number of negative UDS for opiates was adopted as a measure of opiate abuse during and after treatment. Missed urine samples were not taken into account for data analysis.
The number of drug treatments and detoxification and rehabilitation treatments were recorded as this offered another estimate of treatment retention. Attendance was determined as duration from the date of admission to date of the last visit. The number of treatment sessions attended by the patients including the total number of counseling and self-help sessions was calculated. This measure was used to reflect participation in the treatment process. The number of patients who dropped out of treatment was also considered. Attrition was defined as patients who stopped attending the treatment program within 7 days of admission. Attendance at aftercare treatment was also recorded. The opiate-dependent patients (n = 183) were divided into good (n = 66), moderate (n = 32), and poor (n = 85) responders to the biopsychosocial treatment based on the biological, clinical, and psychological outcome measures mentioned above.
SNP selection and genotyping
To date, ten human OPRM1 splice variants have been identified. They contain the same exons 1, 2, and 3 of the normal human OPRM1 (Gene ID: 4988, Gene bank Accession #: NM_000914.2, Gene Alias: KIAA0403, MOR, MOR1, OPRM), which has four exons. However, they differ in their splicing downstream of exon 3. All splice variants result in amino acid sequence changes to the C-terminus of the μ-opioid receptor and may affect the activity of the receptor.Citation47–Citation49 Of these, the normal human OPRM1 gene has been most extensively investigated (). In this study, 22 OPRM1 SNPs were selected from public databases including the following: the SNP database of the National Centre for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/SNP/), the Applied Biosystems SNP database (http://www.appliedbiosystems.com), and the International HapMap Project (http://www.hapmap.org/). The positions of the SNPs in OPRM1 and the relative distance to the translation initiation site are given in .
Table 1 List of SNPs within the OPRM1 gene, their positions, and genotyping data based on the whole cohort (N = 183)
Genomic DNA was extracted from whole blood using a Gentra Puregene Kit (QIAGEN Inc, Valencia, CA). All individuals were genotyped for the chosen 22 SNPs within the OPRM1 gene using the Sequenom MassARRAY® system (iPLEX GOLD) (Sequenom, San Diego, CA). Briefly, PCR and single-base extension primers (SBE) were designed using MassARRAY assay design software (3.1; Sequenom), which allows iPLEX reactions for SBE designs with the modified masses associated with the termination mix. The manufacturer’s instructions for the multiplex reaction were followed throughout the entire process, including the polymerase chain reaction (PCR) amplification (Sequenom), the shrimp alkaline phosphatase (SAP) enzyme (Sequenom) treatment to dephosphorylate the dNTPs unincorporated in the PCR, the SBE reactions using an iPLEX GOLD assay (Sequenom), and the clean-up with a resin kit (Sequenom) to desalt the iPLEX reaction products. PCR and SBE primer sequences and all protocol conditions are available upon request. Reaction products were dispensed onto a 384-element SpectroCHIP bioarray (Sequenom) using a MassARRAY Nanodispenser and assayed on the MassARRAY platform. Mass differences were detected with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). MassARRAY Workstation software (v.3.3; Sequenom) was used to process and analyze the iPLEX SpectroCHIP bioarray. Typer Analyzer v.4.0.2 software (Sequenom) was used to analyze all genotypes obtained from the assays.
Statistical analysis
Consistency with Hardy-Weinberg equilibrium (HWE) was tested using an exact test implemented in PowerMarker software (version 3.25; Paul Lewis, Bioinformatics, North Carolina State University: see http://statgen.ncsu.edu/powermarker).Citation50 Continuous variables were compared using the ANOVA F-test, Kruskal-Wallis test, Wilcoxon rank-sum test, Welch test, and Student’s t-test as appropriate. Categorical variables were analyzed using Pearson’s χ2 tests, and Fisher’s exact test. A significance level of α = 0.05 was applied and a variance of P < 0.05 was considered to be statistically significant. All statistical analyzes were carried out using R software (http://www.r-project.org/).
Results
Sample characteristic
The study group consisted of 183 unrelated Jordanian Arab males meeting the DSM-IV criteriaCitation43 for opiate dependence. The majority of these patients (92%) also had nicotine co-dependence. Cannabis abuse was common (58%) and 53% were alcoholics. The mean age (±SD) of these patients was 33± (8.6) years with a range of 18 to 58 years. The median age of the patients was 32 (range: 18 to 58). Both genotype and phenotype data were available for these patients.
All patients received biopsychosocial treatment at the NCRA and the DRC-PSD for 8 weeks. There were 66 (36%) good responders, 32 (18%) moderate responders, and 85 (46%) poor responders. The description of the three groups, including demographic, nicotine status, opiate consumption (beginning and last taken amount [grams per day]), drug use, dependence variables, substance screening, drug toxicity, psychiatric status, and hospitalization treatment is given in .
Table 2 Details of the 183 opiate-dependent patients classified into three groups
Association of SNP OPRM1 genotypes with opiate-dependence variables and treatment response
Genotypes determined by Sequenom MassARRAY® system for the 22 OPRM1 SNPs were highly accurate with an average success rate of 99.6%. The average genotype discrepancy rate across the 22 loci was only 0.05% (±0.12%) in the samples.
For the 183 opiate-dependent patients, no deviations from HWE were observed for the allele and genotype frequencies for the 22 SNPs (P > 0.5). When comparing the three inclusion groups (good, moderate, and poor responders), significant differences in proportions among genotypes were observed at two sites of the OPRM1 gene with response to the biopsychosocial treatment (rs6912029 [G-172T], P = 0.0329 and rs12205732 [G-1510A], P = 0.0333) (). Specifically, the GG-172 and GG-1510 carriers were more frequent in nonresponders to treatment. For example, the GG-172 carriers were 48% of the nonresponders group (n = 88), whereas they were 18% of the moderate responders group (n = 35) and were 34% of the responders group (n = 70).
Table 4 Association of OPRM1 SNPs with treatment outcome
There were also significant differences in the proportion of opiate use at treatment admission at six sites of the OPRM1 gene (rs2075572 [C644–83G], P = 0.011, and rs648893 [C1165−1189T], P = 0.014, rs609148 [G1165−8803T], P = 0.028, rs9322447 [G1164+11714A], P = 0.038, rs671531 [A1164+28135A G], P = 0.032, rs540825 [T1164+1839A], P = 0.045). However, there were no significant differences for the 22 SNPs for the remaining opiate-dependence variables (age at first use, age at regular use, years of regular use, and frequency of use) excluding the six sites mentioned above concerning the frequency of opiate use ().
Table 3 Differences in genotyping distribution of OPRM1 SNPs with frequency of opiate use
The duration since last positive urine screening for opiates was not significantly different according to the different genotypes (P > 0.3, data not shown). Opiate drug consumption and the number of drug treatments, detoxification, and rehabilitation were also not significantly different among opiate-dependent patients (P > 0.2; data not shown). Exploratory comparisons between SNPs’ OPRM1 and psychiatric status (eg, impulsiveness, depression, anxiety, craving of euphoria, and diminution of attention) at admission and past history as obtained from their medical records (family with history of drug use, overdose toxicity, and suicide attempts) found no significant group differences in opiate-dependent patients (P > 0.1, data not shown).
Discussion
Opiate dependence is a significant public health issue with approximately 10 million people abusing illicit opioids worldwide.Citation51 Hulse et al suggested that opiate dependence is not only associated with high mortality rates and poor health among dependent individuals, but also imposes excessively large economic and social costs upon the community in general.Citation3 Considerable medical, legal, and interpersonal harm, including mortality, is associated with opiate use.Citation3 The major cause of premature death amongst Jordanian opiate users is accidental overdose, along with infectious diseases.Citation52 Moreover, a high prevalence of criminal activity and psychosocial difficulties are also found among Jordanian heroin users.Citation52
The extent of this serious problem has stimulated considerable interest in the physiological and neurochemical processes involved in opioid dependence. In this respect, there is growing evidence that opiate dependence is influenced by genetic factors.Citation53–Citation55 Several studies have been undertaken to estimate the role that genetic factors play in the vulnerability to opiate dependence.Citation19,Citation56–Citation59 These studies have shown that the endogenous opioid system in particular is considered to be one of the most important signaling pathways involved in opiate dependence.Citation60 This system includes the μ opioid receptor, which is the primary site of action for the most commonly used opioids including morphine, heroin, and fentanyl and μ-opioid receptor antagonists such as naltrexone,Citation24 agonists such as methadoneCitation25 or partial antagonists such as buprenorphine.Citation20 This system may also play a crucial role in mediating the reinforcement effects of nonopioid drugs such as alcohol, cannabinoids, nicotine, and cocaine.Citation60–Citation62
In the current study, our aim was to identify the genetic factors associated with responsiveness to the biopsychosocial treatment offered for opiate drug-dependent patients at the NCRA and the DRC-PSD. Drug-dependent patients of Arab descent were genotyped for variance in the OPRM1 gene. The genotyping was carried out by sequenom MassARRAY® system for 22 OPRM1 SNPs. All OPRM1 polymorphisms, which were genotyped in this study, are reported in the NCBI database (http://www.ncbi.nlm.nih.gov/SNP/). This is, to our knowledge, the first attempt to evaluate a series of SNPs spanning the coding sequence of the OPRM1 gene in an opiate-using Jordanian Arab population in relation to the response to the biopsychosocial treatment.
Various alcohol or drug dependence-related association studies have expanded their investigation to include up to 20 SNPs spanning the coding sequence of the OPRM1 gene; all include the A118G (Asn40Asp) variants. For instance, Japanese subjects meeting ICD-10 criteria for methamphetamine (MAP) dependence and controls were genotyped for 20 SNPs including 10 SNPs in the 3′-UTR region.Citation63 The study reported that A118G and other SNPs were not associated with MAP dependence or psychosis, and the rs2075572 G-allele was significantly associated only with increased risk for a diagnosis of MAP dependence or psychosis (P = 0.011). Ten SNPs selected throughout OPRM1 were examined within a Chinese population to investigate the relationship between the SNPs and heroin-induced subjective responses.Citation64 The study reported that three SNPs in intron 1 were associated with an increased risk of positive responses on first use of heroin and were likely to contribute to further heroin consumption; A118G and rs2075572 were not associated with any differences in heroin-induced subjective responses. In another association study, eight SNPs within OPRM1 in alcohol-l, cocaine-, opioid-, and polysubstance-dependent European Americans (EA) and African Americans (AA) were genotyped.Citation65 The EA and AA study reported that C-2044A polymorphism was associated with the combination of alcohol and opioid dependence in EA subjects, but not AA subjects. Again, A118G was not associated with any of the substance-dependent phenotypes. A genetic association study on the role of OPRM1 genetic variations in a large case-control sample of alcohol- and drug- (cocaine and opioid) dependent European Americans was conducted by Zhang et al.Citation38 They typed 13 SNPs representing the major haplotypes observed in HapMap, all of which are included in the present study. They found that seven SNPs (but not rs1799971 [A188G]) were associated with alcohol, opioid, and cocaine dependency. Zhang et alCitation38 found that the frequency of the rs524731A and rs648893 T alleles was significantly higher among EA than AA subjects. Finally, a case-control study of opiate and nonopiate-dependent Jordanian Arabs was recently conducted by AL-Eitan et alCitation66 to investigate the genetic association of 22 SNPs spanning the coding sequence of the OPRM1 locus with opiate dependence.Citation66 The study reported that three SNPs (rs6912029 [G-172T], rs12205732 [G-1510A], and rs563649 [G-983A]) were associated with opiate dependence.Citation66 In the present study, these 22 SNPs were also tested for a possible association of OPRM1 polymorphisms with response to the biopsychosocial treatment.
Analysis of the relationship between treatment response and the OPRM1 SNP genotypes showed there was a significant difference in the genotyping distribution between the three inclusion groups at two sites (rs6912029 [G-172T], and rs12205732 [G-1510A]) of the OPRM1 gene located in the upstream (5′-UTR) region. UTRs are known to play crucial roles in the post-transcriptional regulation of gene expression, including modulation of the transport of mRNAs out of the nucleus and of translation efficiency,Citation67 subcellular localization,Citation68 and stability.Citation69 The importance of UTRs in regulating gene expression is underlined by the finding that mutations that alter the UTR can lead to serious pathology.
Previous studies have reported an association of the OPRM1 118A > G with different opioidsCitation22,Citation70–Citation73 and opiate antagonist “naltrexone” treatment.Citation37,Citation72 However, we found no influence of this SNP on responsiveness to treatment, as the allele and genotypes’ frequencies were similar in the opiate-dependent patients with a P > 0.05. A recent study demonstrated that individuals with the Asp40 variants of the OPRM1 gene showed favorably higher relapse prevention rates when receiving naltrexone treatment.Citation42 A recent clinical trial also showed that the variants of the OPRM1 gene did not have any preferential effect on naltrexone treatment in alcoholics.Citation70 The functional importance of treatment on any of the variants of the OPRM1 gene is still being elucidated. Although earlier studies in transfected cells reported that the OPRM1-Asp40 (118G) variant had a threefold higher affinity for β-endorphin than OPRM1-Asn40 (G118), which would suggest enhanced function,Citation23 this was not reported by others.Citation33,Citation34 In addition, recent in vitro transfection studies have suggested that the Asp40 allele might be associated with lower OPRM1 protein expression than the Asp40 allele.Citation35 As a result, a higher frequency of this allele would have been more common in the individuals with poor treatment response.
Various genetic studies have reported that the frequency of the Asp40 allele can vary significantly between populations with drug dependence from as low as 5% in African AmericansCitation15 to 16% in European AmericansCitation38 to 26% in this study of Arab descentCitation66 and as high as 58% among those of Asian descent.Citation39 Although the complexity of opiate dependence and the differences in ethnicity have influences on the OPRM1 results, all the previous studies have confidence in the hypothesis that the Asp40 allele might be associated with lower OPRM1 protein expression than the Asn40 allele.Citation35 As a result, a higher frequency of the Asp40 allele would have been expected in the poor responder group as compared to the responder group. Similarly, this study found that opiate-dependent patients with poor treatment response had a higher frequency of Asp40 allele and this was in agreement with the proposed theory.
In a search for the relationship between the dependence variables of opiate dependence and the OPRM1 SNPs’ genotypes, none of the analyzed SNPs in this study were associated with the age of opiate initiation, transition to opiate dependence, and regular use and opiate consumption (beginning and last taken amount [grams per day]). However, the data indicated that there were significant differences in opiate frequency use at only six sites (rs2075572 [C644−83G], rs648893 [C1165−1189T], rs609148 [G1165−8803T], rs671531 [A1164+28135A G], rs9322447 [G1164+11714A], rs540825 [T1164+1839A]) of the OPRM1 gene with P < 0.05. Previous studies have indicated that OPRM1 polymorphism may be associated with clinical variables such as treatment duration, hospitalization for drug treatments, detoxification, rehabilitation, counseling and self-help for substance abuse, psychiatric symptoms, history of drug use, overdose toxicity, and follow-up measures (urine screening test).Citation15,Citation39,Citation45,Citation46,Citation62,Citation71 Unfortunately, the P values concerning the clinical variables in this study were not significant.
This discrepancy between the study results could be related to the past history, the psychiatric status, and the subgroups of drug-dependent individuals with violent behavior. This might be due to a bias in classifying the drug-dependent individuals. There are some possible confounding factors that should be taken into consideration when assessing the patients with drug dependence such as the methods used for defining violent, antisocial behavior and the early onset of dependence. Several studies have based patients’ assessment on collecting the phenotypic data by only a self-report, interviewing the patients by health professional workers, or obtaining the data from their medical records, including this study.
Previous studies have shown that some OPRM1 markers deviate from the Hardy-Weinberg Equilibrium (HWE) in different populations.Citation38,Citation74 There are many possible confounding factors for this deviation including population stratification, genotyping errors, and true association with phenotypes.Citation74 In this study, the genotypic frequencies of the OPRM1 markers met HWE expectations. Other studies have also reported sample bias or genotyping errors. Our patients were from one geographic origin and were 100% native Jordanians of Arab ancestry, which is a population known to be genetically homogenous. Genotyping errors were minimized by genotyping each patient twice in order to avoid technical errors as evidenced by the low average rate of genotype discrepancy. SNPs’ genotyping was conducted for patients under the same conditions and during the same period. Genotypes were also evaluated by investigators, who were blind to the status of the subject and any discrepancies were resolved by test replication. Finally, only male individuals with opiate dependence were studied. Therefore, the generalization of the results to all drug-dependent patients is limited.
In conclusion, this is the first report of an association between the OPRM1 G-172T and G-1510A polymorphisms and response to the biopsychosocial treatment. Specifically, the GG-172 and GG-1510 carriers were more frequent in nonresponders to treatment. This may lead to more accurate matching of individuals to different treatment options and early identification of persons at high risk of relapse and therefore requiring more intensive intervention. However, further pharmacogenetic studies are needed to confirm the present findings on the influence of the OPRM1 G-172T and G-1510A on the response to treatment in opiate-dependent patients of Arab descent.
Acknowledgements
Publication number LA011-007 of the Centre for Forensic Science at the University of Western Australia. We gratefully acknowledge the contribution of participating patients whose cooperation made this study possible. Funding for this project was provided in part by Centre for Forensic Science and Unit for Research and Education in Alcohol and Drugs of the School of Psychiatry and Clinical Neurosciences, The University of Western Australia.
Disclosure
The authors report no conflicts of interest in this work.
References
- TsuangMTLyonsMJMeyerJMCo-occurrence of abuse of different drugs in men: the role of drugs-specific and shared vulnerabilitiesArch Gen Psychiatry1998559679729819064
- AnthonyJCWarnerLAKesslerRCComparative epidemiology of dependence on tobacco, alcohol, controlled substances, and inhalants: basic findings from the national comorbidity surveyExp Clin Psychopharmacol19942244268
- HulseGKEnglishDRMilneEThe quantification of mortality resulting from the regular use of illicit opiatesAddiction19999422122910396790
- NuttDKingLASaulsburyWBlakemoreCDevelopment of a rational scale to assess the harm of drugs of potential misuseLancet20073691047106317382831
- BamentDCookeRWeekleyJAliRTreatment outcomes at 12 months post admission to drug treatment: The third report of the South Australian component of the Australian Treatment Outcomes Study – Heroin Adelaide: DASC Monograph No 16 Research Series2004
- BallJRossAThe Effectiveness of Methadone Maintenance TreatmentNew York, NYSpringer-Verlag1991
- JohnsonBAAddiction Medicine: Science and PracticeNew York, NYSpringer2011
- EapCBBuclinTBaumannPInterindividual variability of the clinical pharmacokinetics of methadone: implications for the treatment of opioid dependenceClin Pharmacokinet2002411153119312405865
- CrettolSDéglonJJBessonJABCB1 and cytochrome P450 genotypes and phenotypes: influence on methadone plasma levels and response to treatmentClin Pharmacol Ther20068066868117178267
- SrisurapanontMJarusuraisinNNaltrexone for the treatment of alcoholism: a meta-analysis of randomized controlled trialsInt J Neuropsychopharmacol20058226728015850502
- StreetonCWhelanGNaltrexone, a relapse prevention maintenance treatment of alcohol dependence: a meta-analysis of randomized controlled trialsAlcohol2001366544552
- KranzlerHRVan KirkJEfficacy of naltrexone and acamprosate for alcoholism treatment: a meta-analysisAlcohol Clin Exp Res20012591335134111584154
- BouzaCAngelesMMunozAAmateJEfficacy and safety of naltrexone and acamprosate in the treatment of alcohol dependence: a systematic reviewAddiction200499781182815200577
- AntonRFO'MalleySSCirauloDACOMBINE Study Research Group. Combined pharmacotherapies and behavioral interventions for alcohol dependence: the COMBINE study: a randomized controlled trialJAMA2006295172003201716670409
- GelernterJKranzlerHCubellsJGenetics of two mu opioid receptor gene (OPRM1) exon I polymorphisms: population studies, and allele frequencies in alcohol- and drug-dependent subjectsMol Psychiatry1999447648310523821
- AntonRFOrosziGO'MalleySAn evaluation of mu opioid receptor (OPRM1) as a predictor of naltrexone response in the treatment of alcohol dependence: results from the combined pharmacotherapies and behavioral interventions for alcohol dependence (COMBINE) studyArch Gen Psychiatry200865213514418250251
- KimSGKimCMKangDHAssociation of functional opioid receptor genotypes with alcohol dependence in KoreansAlcohol Clin Exp Res20042898699015252283
- GianoulakisCKrishnanBThavundayilJEnhanced sensitivity of pituitary β-endorphin to ethanol in subjects at high risk of alcoholismArch Gen Psychiatry19965332502578611062
- HaileCNKostenTAKostenTRPharmacogenetic treatments for drug addiction: alcohol and opiatesAm J Drug Alcohol Abuse200834435538118584566
- KreekMJVocciFJHistory and current status of opioid maintenance treatments: blending conference sessionJ Subst Abuse Treat20022329310512220607
- KreekMJBartGLillyCPharmacogenetics and human molecular genetics of opiate and cocaine addictions and their treatmentsPharmacol2005571126
- RavindranathanAJoslynGRobertsonMFunctional characterization of human variants of the mu opioid receptor geneProc Natl Acad Sci U S A200910626108111081619528663
- BeyerAKochTSchröderHSchulzSHolltVEffect of the A118G polymorphism on binding affinity, potency and agonist-mediated endocytosis, desensitization, and resensitization of the human mu opioid receptorJ Neurochem20048955356015086512
- VolpicelliJRAltermanAIHayashidaMO'BrienCPNaltrexone in the treatment of alcohol dependenceArch Gen Psychiatry199249118768801345133
- DoleVPNyswanderMA medical treatment for diacetylmorphine (heroin) addiction. A clinical trial with methadone hydrochlorideJAMA196519364665014321530
- BenyheSTóthGKeveiJCharacterization of rat brain opioid receptors by (Tyr-3, 5-3H)1,D-Ala2,Leu5-enkephalin bindingNeurochem Res1985106276352989719
- ChenYMestekALiuJHurleyJAYuLMolecular cloning and functional expression of a μ-opioid receptor from rat brainMol Pharmacol1993448128393525
- DelfsCLKongHMestekAExpression of mu opioid receptor mRNA in rat brain: an in situ hybridization study at single cell levelJ Comp Neurol199434546688089277
- WaldhoerMBartlettSEWhistlerJLOpioid receptorsAnnu Rev Biochem20047395399015189164
- HoeheMRKopkeKWendelBSequence variability and candidate gene analysis in complex disease: association of mu opioid receptor gene variation with substance dependenceHum Mol Gene2000928952908
- IkedaMRIdeSHanWHow individual sensitivity to opiates can be predicted by gene analysesTrend Pharmacol Sci200526311317
- BergenAWKoskozkaJPetersonRMu opioid receptor gene variants: lack of association with alcohol dependenceMol Psychiatry199724904979399694
- BondCLaforgeKSTianMSingle nucleotide polymorphism in the human mu opioid receptor gene alters beta-endorphin binding and activity: possible implications for opiate addictionProc Natl Acad Sci U S A19989516960896139689128
- BefortKFilliolDDécaillotFMSingle nucleotide polymorphic mutation in the human mu opioid receptor severely impairs receptor signallingJ Biol Chem20012763130313711067846
- ZhangYWangDJohnsonADPappACSadeeWAllelic expression imbalance of human mu opioid receptor (OPRM1) caused by variant A118GJ Biol Chem200528038326183262416046395
- AriasAFeinnRKranzlerHRAssociation of an Asn40Asp (A118G) polymorphism in the mu opioid receptor gene with substance dependence: a meta-analysisDrug and Alcohol Depend200683262268
- OrosziGAntonRFO'MalleySOPRM1 Asn40Asp predicts response to naltrexone treatment: A haplotype based approachExp Clin Psychopharmacol2009333383393
- ZhangHLuoXKranzlerHRAssociation between two mu opioid receptor gene (OPRM1) haplotype blocks and drug or alcohol dependenceHum Mol Genet20061580781916476706
- KimSGKimCMChoiSWA mu opioid receptor gene polymorphism (A118G) and naltrexone treatment response in adherent Korean alcohol-dependent patientsPsychopharmacology200920161161818795264
- KarhuvaaraSSimojokiKVirtaATargeted nalmefene with simple medical management in the treatment of heavy drinkers: a randomized double-blind placebo-controlled multicentre studyAlcohol Clin Exp Res2007311179118717451401
- KrystalJHCramerJAKrolWFKirkGFRosenheckRANaltrexone in the treatment of alcohol dependenceN Engl J Med20013451734173911742047
- OslinDWBerrettiniWKranzlerHRA functional polymorphism of the mu opioid receptor gene is associated with naltrexone response in alcohol-dependent patientsNeuropsychopharmacology2003281546155212813472
- American Psychiatric AssociationDiagnostic and Statistical Manual of Mental Disorders4th edWashington, D.C.American Psychiatric Association1994
- McLellanATCacciolaJKushnerHThe fifth edition of the addiction severity index: Cautions, additions and normative dataJ Subst Abuse Treat19929461480
- McGueMPickensRWSvikisDSSex and age effects on the inheritance of alcohol problems: a twin studyJ Abnorm Psychol19921013171537970
- BerrettiniWHPersicoAMDopamine D2 receptor gene polymorphisms and vulnerability to substance abuse in African-AmericansBiol Psychiatry1996401441478793046
- BareLAManssonEYangDExpression of two variants of the human mu opioid receptor mRNA in SK-N-SH cells and human brainFEBS Lett199435422132167957926
- XinLWangZJBioinformatics analysis of the human μ opioid receptor (OPRM1) splice and polymorphic variantsAAPS Pharm Sci20024113
- PasternakGWMultiple opiate receptors: déjà vu all over againNeuropharmacology2004131232315464147
- LiuKMuseSVPowerMarker: integrated analysis environment for genetic marker dataBioinformatics2005212128212915705655
- United Nations Office on Drug and Crime (UNODC), country profile, The Hashemite Kingdom of Jordan2010Vienna, Austria Available from: http://www.unodc.org/egypt/en/country_profile_jordan.html. Accessed December 11, 2011
- HadidiMSIbrahimMIAbdallatIMCurrent trends in drug abuse-associated fatalities – Jordan, 2000–2004Forensic Sci Int2009186444719217732
- Di ChiaraGImperatoADrugs abused by humans preferentially increase synaptic dopamine concentrations in the mesolimbic system of freely moving ratsProc Natl Acad Sci U S A199885527452782899326
- TandaGPontieriFEDi ChiaraGCannabinoid and heroin activation of mesolimbic dopamine transmission by a common m1 opioid receptor mechanismScience1997276204820509197269
- WeissFLorangMTBloomFEKoobGFOral alcohol self-administration stimulates dopamine release in the rat nucleus accumbens. Genetic and motivational determinantsJ Pharmacol Exp Ther19932672502588229752
- NestlerEJGenes and addictionNat Genet200026227281
- NestlerEJMolecular neurobiology of addictionAm J Addict20011020121711579619
- NestlerEJOpportunities for understanding addictionJ Neurosci2001218324832711606619
- NestlerEJIs there a common molecular pathway for addiction?Nat Neurosci200581445144916251986
- HerzAEndogenous opioid systems and alcohol addictionPsychopharmacology1997129991119040115
- LaforgeKSShickVSpanglerRDetection of single nucleotide polymorphisms of the human mu opioid receptor gene by hybridization or single nucleotide extension on custom oligonucleotide gelpad microchips: potential in studies of addictionAm J Med Genet20009660461511054767
- EhlersCLLindPAWilhelmsenKCAssociation between single nucleotide polymorphisms in the mu opioid receptor gene (OPRM1) and self-reported responses to alcohol in American IndiansBio Med Central Medical Genetics2008935
- IdeSKobayashiHTanakaKGene polymorphisms of the mu opioid receptor in methamphetamine abusersAnn N Y Acad Sci2004102531632415542732
- ZhangDShaoCShaoMEffect of mu opioid receptor gene polymorphisms on heroin-induced subjective responses in a Chinese populationBiol Psychiatry2007611244125117157823
- LuoXKranzlerHRZhaoHGelernterJHaplotypes at the OPRM1 locus are associated with susceptibility to substance dependence in European-AmericansAm J Med Genet B Neuropsychiatr Genet200312019710812815747
- AL-EitanLNJaradatADadourRTayGKHulseGKPolymorphisms in the μ-opioid receptor gene in Jordanian Arabs with opiate drug dependenceScience MED201239197
- Van der VeldenAWThomasAAThe role of the 5′ untranslated region of an mRNA in translation regulation during developmentInt J Biochem Cell Biol1999318710610216946
- JansenRPmRNA localization: message on the moveNat Rev Mol Cell Biol2001224725611283722
- BashirullahACooperstockRLLipshitzHDSpatial and temporal control of RNA stabilityProc Natl Acad Sci U S A2001987025702811416182
- LötschJZimmermannMDarimontJDoes the A118G polymorphism at the mu opioid receptor gene protect against morphine-6-glucuronide toxicity?Anesthesiology20029781481912357145
- OertelBGSchmidtRSchneiderAGeisslingerGLotschJThe mu opioid receptor gene polymorphism 118A > G depletes alfentanil-induced analgesia and protects against respiratory depression in homozygous carriersPharmacogenet Genomics20061662563616906017
- RombergRROlofsenEBijlHPolymorphism of mu opioid receptor gene (OPRM1:c.118A > G) does not protect against opioid-induced respiratory depression despite reduced analgesic responseAnesthesiology200510252253015731588
- CrettolSBessonJCroquette-KrokarMAssociation of dopamine and opioid receptor genetic polymorphisms with response to methadone maintenance treatmentProg Neuropsychopharmacol Biol Psychiatry2008321722172718687376
- GuoSWThompsonEAPerforming the exact test of Hardy-Weinberg proportion for multiple allelesBiometrics1992483613371637966