Abstract
The objective of this study was to determine copy number variant (CNV) and promoter genetic variants in glutathione S-transferase Mu class 1 (GSTM1) and the risk of recurrence (REC)/second primary tumor (SPT) in patients with previously diagnosed early stage head and neck cancer. Among 441 subjects, 133 experienced REC and/or an SPT, while 308 had single primary disease. TaqMan real-time polymerase chain reaction was used to measure the exact copy number of GSTM1 and direct sequencing was used to determine genetic variants in the GSTM1 promoter region. Multivariate Cox regression analysis was performed to estimate hazard ratios (HRs) and 95% confidence intervals (95% CIs) associated with copy number and genetic variants. REC/SPT-free survival times were compared by constructing Kaplan–Meier curves and differences between curves were tested by logrank test. Results showed a significantly decreased REC/SPT (HR = 0.57; 95% CI = 0.35–0.95) and longer REC/SPT-free survival in subjects with at least two copies of GSTM1 compared with the GSTM1 homozygous deletion, but not in those with one copy of GSTM1. The −498G, −426G, and −339T alleles were significantly associated with REC/SPT, with HRs of 0.11 (0.02–0.85), 0.28 (0.11–0.74) and 2.02 (1.07–3.82), respectively. Kaplan–Meier survival analysis showed that the −498G, −426G, and −339C alleles were also significantly associated with increased REC/SPT-free survival. Further haplotype analysis showed the haplotype P−498G-−426G-−339C carriers had decreased REC/SPT with a HR of 0.09 (95% CI 0.01–0.71) and increased REC/SPT-free survival compared with those with haplotype P−498C-−426A-−339T. The P−498C-−426A-−339T-containing reporter construct had significantly increased luciferase expression. These results suggest that the GSTM1 CNV and promoter haplotype are better predictors of REC/SPTs of head and neck cancer than just measuring the presence/absence of GSTM1.
Introduction
Head and neck cancer (HNC), which includes carcinomas of the oral cavity, pharynx, and larynx, is one of the most common human cancers worldwide. HNC accounts for 3%–5% of all cancers in the USA and it has been estimated that in 2012 more than 52,000 (40,250 oral and pharynx and 12,360 larynx) individuals will be diagnosed with HNC and 11,500 (7850 oral and pharynx and 3650 larynx) will die of the disease.Citation1 Treatment of HNC is limited in early stage disease to either radiotherapy or surgery.Citation2 However, post-stage treatment morbidity and mortality increases when patients experience recurrence of disease (REC) or develop a second primary tumor (SPT).Citation3 SPTs and recurrences develop in up to 20% of patients within 5 years of curative treatment.Citation4 Smoking and alcohol consumption are strongly associated with poor outcomes in HNC.Citation5,Citation6 In addition, inherited factors, including genetic variants at specific genes, have also been demonstrated to modify this risk.Citation7
Glutathione S-transferase Mu (GSTM) class enzymes, members of the glutathione S-transferase (GST) superfamily of phase II drug-metabolizing enzymes, play an important role in protecting cells against xenobiotics by conjugating with glutathione to detoxify electrophilic compounds. The genes encoding the GSTM family are located on chromosome 1p13.3 in the order GSTM4, GSTM2, GSTM1, GSTM5, GSTM3.Citation8,Citation9 GSTM proteins have distinct tissue distribution: GSTM1 is a major Mu-class GST in the liver, the GSTM2 subunit is primarily associated with skeletal muscle, GSTM3 is enriched in testis, and GSTM5 is found in brain.Citation10 It is well known that hepatic GSTM1 is highly polymorphic, and these genetic variations, which include copy number variants (CNVs) and common single nucleotide polymorphisms (SNPs), are likely to contribute to inter-individual differences in response to carcinogens and drugs. The homozygous deletion of the GSTM1 gene, which results in the absence of the GSTM1 enzyme, is present in 48%–57% of Caucasians, 23%–41% of African Americans, 32%–53% of Asians, and 40%–53% of Hispanics.Citation11 Due to the important role of GSTM1 in the metabolism of carcinogens and drugs, this deletion variant in GSTM1 has been demonstrated to contribute to cancer susceptibility as well as to the prognosis of certain cancers.Citation12–Citation17 Previous studies have assessed risk based on the presence or absence of the GSTM1 gene, without determining the exact number of gene copies in individuals who possess GSTM1. Furthermore, potential functional promoter SNPs in the GSTM1 gene could influence the protein expression level and thus change the response of cells to exogenous carcinogens and drugs. However, investigation into these promoter genetic variants is still limited.
In this study, we report the results of the association between exact CNVs and promoter SNPs in GSTM1 with HNC REC/SPT. Our results indicate that this approach to assessing genetic variation within GSTM1 is a more superior prognostic biomarker than just determining the presence/absence of the gene and will aid in the identification of high-risk/poor-outcome individuals. Future replication studies in other independent cohorts are warranted to confirm these findings.
Methods
Human liver tissues and GST expression detection
A total of 111 human liver tissue samples were obtained from the US Cooperative Tissue Network (Birmingham, AL, USA) and liver cytosols were prepared by 100,000 g centrifugation of tissue homogenized in 20 mM Tris-hydrochloric acid (pH 7.8)-buffered 0.25 M sucrose containing 0.5 mM ethylenediaminetetraacetic acid, 20 μM butylated hydroxytoluene, and 0.1 mM dithiothreitol, as previously described.Citation18 GSTs were determined after glutathione-agarose affinity chromatography and wide-pore high-performance liquid chromatography, as described by Coles and Kadlubar.Citation19
Study subjects
The study population has been described previously.Citation4,Citation20–Citation23 Briefly, patients included in this study were stage I and II head and neck cases enrolled in the randomized Retinoid Head and Neck Second Primary Trial from 1991 to 1999 at MD Anderson Cancer Center, in which patients either received daily low dose (30 mg/day) of 13-cis-retinoic acid or placebo for 3 years. Patients must have remained cancer free for at least 16 weeks following surgery and/or radiation treatment to be enrolled in the trial. There were no recruitment restrictions on age, gender, or ethnicity. Before randomization, patients were given a structured questionnaire that elicited information on sociodemographic factors, tobacco-use history, alcohol consumption, and other exposures. Clinical data were obtained by medical chart review. The definitions of “second primary tumor” and “recurrence” following the Warren and Gates criteria were provided previously.Citation4
This study included 441 head and neck patients. Among these cases, 133 experienced recurrence of disease and/or an SPT, while 308 had single primary disease. All patients signed written informed consent before participation in the study and the study was approved by the Institution Review Board of the University of Texas MD Anderson Cancer Center. “Never smokers” were individuals who had smoked less than 100 cigarettes during their lifetime. “Former smokers” were individuals who had stopped smoking for at least a year at the time of enrollment.
Determination of GSTM1 copy number
The gene copy number of GSTM1 within the genome was determined with a TaqMan copy number assay (Applied Biosystems, Foster City, CA, USA), which used genomic DNA as a template and ran as a triplex TaqMan real-time polymerase chain reaction (PCR) with Rnase P as the reference gene, along with a well-characterized reference sample (Coriell Institute for Medical Research, Camden, NJ, USA) with two copies of GSTM1 as a calibrator. Real-time PCR data were analyzed by the comparative Ct method to calculate relative changes in the copy number of GSTM1. All CNV genotypes were determined without knowledge of the REC/SPT status of the subjects.
Determination of genotypes in the promoter of GSTM1
GSTM1 promoter fragments from each individual were amplified using the forward primer (GSTM1-PF: 5′-CAG GTT GGA CAT TGT TCT CGT G-3′) and reverse primer (GSTM1-PR: 5′-CAG CTG CTT CGC ACT TCC CT-3′) to produce a 1924 bp fragment. Genetic variants were identified by direct sequencing of the PCR products with a GenomeLab DTCS Quick Start Kit in a Beckman GeXP Genome Analyzer (both from Beckman Coulter, Fullerton, CA, USA). The sequencing primers were GSTM1-seq1 (5′-GGA GTT TCT TCA GAC TCA CAA T-3′), GSTM1-seq2 (5′-CCT GGG CCT TAA AGC ATG AC-3′), and GSTM1-seq3 (5′-CAC AGA CCA CAT TTC CTT TAC-3′). Genetic variants were identified using Codon-Code Aligner software (CodonCode, Dedham, MA, USA), a program for sequence assembly and mutation detection.
Construction of reporter plasmids
To verify whether identified SNPs influenced the transcriptional activity of GSTM1, we constructed eight reporter plasmids encompassing base pairs from −1687 to +87 of the human GSTM1 gene promoter. The primers used to amplify the fragment were 5′-GAC TAC GCG TTA CTG AAG AAC ACA CAT GG-3′ and 5′-GAA TAG ATC TGC GGA TGT CCC AGT ACC-3′, which contain Mlu I and Bgl II restriction sites (the underlined sequences), respectively. After amplification using LA Taq polymerase (NEB, Ipswich, MA, USA), the PCR product with the GSTM1 promoter was digested with Mlu I and Bgl II then inserted into a pGL3-basic vector (Promega, Madison, WI, USA) with the firefly luciferase gene as a reporter. The resulting construct was designated “pC-A-C” (with −498C, −426A, −339C) after sequence verification. This P−498C–426A–339C construct was subsequently used as a template to generate seven other constructs containing all possible haplotypes (P−498C-−426A-−339T, P−498C-−426G-−339C, P−498C-−426G-−339T, P−498G-−426A-−339C, P−498G-−426A-−339T, P−498G-−426G-−339C, P−498G-−426G-−339T) using the Quickchange Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA). All constructs were confirmed by direct sequencing using the Beckman GeXP Genome Analyzer.
Cell culture and luciferase assay
The squamous cell carcinoma oral cavity cell line T409 was cultured in Dulbecco’s modified Eagle’s medium/F12 medium supplemented with 10% fetal bovine serum, 2 mmol/L L-glutamine, 100 U/mL penicillin, and 100 U/mL streptomycin (Invitrogen, Carlsbad, CA, USA) in a humidified, 5% CO2 incubator at 37°C. For transient transfection, 10 × 104 cells were plated in a 96-well plate and grown to 70%–80% confluence. Lipofectamine™ 2000 (Invitrogen) was used to transfect GSTM1-pGL3 basic constructs and control plasmid pRL-SV40 into cells according to the manufacturer’s protocol. Luciferase activity was measured using a TD20/20 luminometer (Turner Designs, Sunnyvale, CA, USA). The empty pGL3-basic vector was also transfected into cells as a control. Fold increase was calculated by defining the activity of empty pGL3 basic vector as 1. Differences were determined by t-test and a P value of <0.05 was considered significant.
Statistical methods
Multivariate Cox regression was used to estimate hazard ratios (HRs) associated with CNV, genotypes, or haplotypes along with 95% confidence intervals (CIs), while adjusting for confounding variables such as age, gender, smoking status, alcohol consumption, tumor site (larynx, oral cavity, and pharynx), stage (I or II), and randomization (13-cis-retinoic acid chemoprevention or placebo). Kaplan–Meier curves were constructed to compare event-free survival by CNV, genotypes, or haplotypes and logrank tests were performed to compare differences between survival curves. All analyses were performed using the Intercooled Stata statistical software package (v 10.0; Stata, College Station, TX, USA). All statistical tests were two sided and a P value of 0.05 was used as the criterion of statistical significance.
Results
GSTM1 protein expression GSTM1 genetic variants
To verify the relationship between CNV and GSTM1 expression, we tested 111 human liver samples. Of these, 50 (45.0%) subjects were without GSTM1 expression and 61 subjects (55.0%) expressed GSTM1 with about 145-fold GSTM1 expression variation.
The results also showed that subjects with at least two copies have much higher GSTM1 expression than those with only one copy number (0.339 vs 0.118, P < 0.001) (). This could partially account for GSTM1 expression variants among individuals, but among carriers with one copy of GSTM1, there was still an 80-fold variation in protein expression. Thus, we conducted resequencing of a 1924-bp section of the 5’ promoter region and found multiple genetic variants (). However, due to the very small sample size, there was no significant correlation between the genetic variants and GSTM1 protein levels.
Figure 1 Level of GSTM1 expression in human liver tissue. (A) Samples have been arranged in order of increasing expression. (B) levels of GSTM1 protein expression in liver tissue by GSTM1 copy number.
Abbreviations: CNV, copy number variant; GST, glutathione S-transferase; GSTM1, glutathione S-transferase Mu class 1.
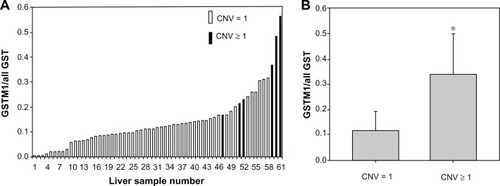
Table 1 List of genetic variants in the promoter of GSTM1 identified in Caucasians
Genetic variation screening
Through TaqMan GSTM1 CNV assay, we determined that 57% of the study subjects had the GSTM1 homozygous deletion, 24% had one copy of GSTM1, and 19% had at least two copies of GSTM1. Of those with the GSTM1 gene, we sequenced the full-length promoter region of GSTM1 and identified one insert variant and 14 SNPs (). Among these, the −888A>T and −341C>T were novel whereas the other SNPs had been previously deposited in the National Center for Biotechnology Information database (−1543 TTCT insertion [rs71794573], −1529C>G [rs36210087], −1490A>G [rs36209763], −1143A>G [rs36209754], −498G>C [rs412543], −486C>G [rs3815029], −471C>T [rs55791819], −426G>A [rs412302], −344C>T [rs4147561], −343A>T [rs4147562], −339C>T [rs4147563], −304G>A [rs28549287], −164 C>T [rs36208869]). The allele frequencies for these variants range from 0.07 to 0.47 ().
CNV of GSTM1 and REC/SPT-free survival of HNC
Regression analysis revealed significantly decreased REC/SPT in subjects with at least two copies of GSTM1 (odds ratio (OR) = 0.57; 95% CI = 0.35–0.95) but not in those with one copy of the gene (). Kaplan–Meier survival analysis revealed longer REC/SPT-free survival in patients with at least two copies of GSTM1. However, there was no significant difference between GSTM1 null subjects and one-copy carriers (). SNP analysis showed the G allele of −498C>G, the G allele of −426A>G, and the T allele of −339C>T SNPs were significantly associated with REC/SPT in HNC patients with HR 0.11 (0.02–0.85), 0.28 (0.11–0.74), and 2.02 (1.07–3.82), respectively (). Kaplan–Meier survival analysis also demonstrated that the −498G, −426G, and −339C alleles were significantly associated with longer REC/SPT-free survival times in HNC subjects ().
Figure 2 Kaplan–Meier plots of time (months) and REC/SPT-free survival in HNC patients according to GSTM1 CNV.
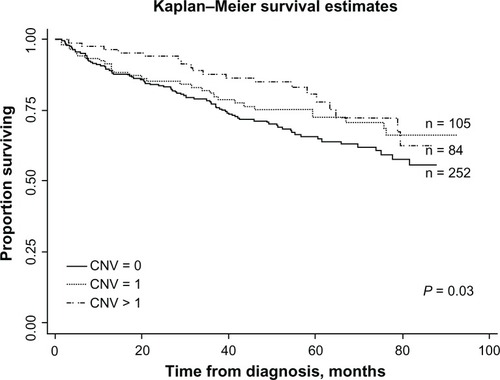
Figure 3 Kaplan–Meier estimates of time (months) and REC/SPT-free survival in HNC patients by GSTM1 promoter SNPs. (A) GSTM1–498C>G SNP and REC/SPT-free survival. (B) GSTM1 −426A>G SNP and REC/SPT-free survival. (C) GSTM1–339C>T SNP and REC/SPT-free survival.
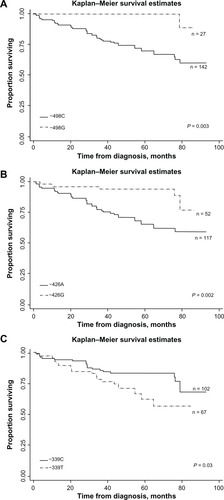
Table 2 Copy number frequencies of GSTM1 and risk of SPT/REC in head and neck cancer
Table 3 Allele frequencies of GSTM1 promoter SNPs and the risk of SPT/REC in head and neck cancer
Haplotypes of GSTM1 promoter variants and REC/SPT-free survival of HNC
Three SNPs (−498C>G, −426A>G, and −339C>T) in the promoter of GSTM1 were independently associated with REC/SPT in HNC patients and were used for the construction of haplotypes. We evaluated the influence of different haplotypes on REC/SPT in our study population. The data showed that the patients with haplotype P−498G-−426G-−339C had decreased REC/SPT (HR = 0.09, 95% CI 0.01–0.71) compared were those with haplotype P−498C-−426A-−339T (). There was no significant difference between other haplotypes and haplotype P−498C-−426A-−339T (). Kaplan–Meier survival estimates showed increased REC/SPT in haplotype P−498G-−426G-−339C group when compared with haplotype P−498C-−426A-−339T group ().
Figure 4 Kaplan–Meier estimates of time (months) and REC/SPT-free survival in HNC patients by in haplotype P-498G−-426G−-339C and P498C−-426A−-339T.
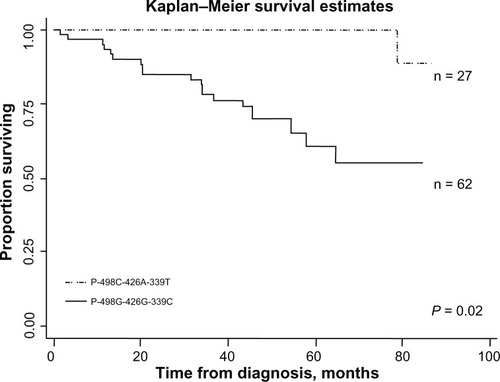
Table 4 GSTM1 haplotype frequencies and risk of SPT/REC in head and neck cancer
Effect of GSTM1 haplotypes on transcriptional activity
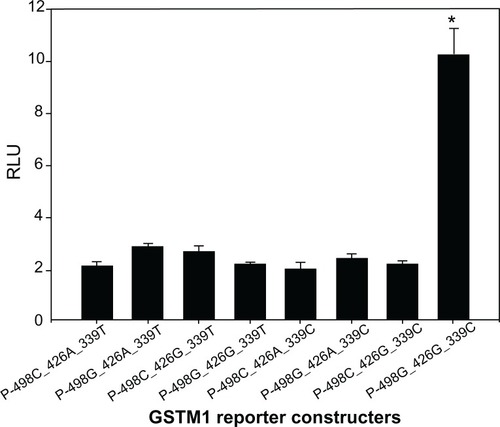
To evaluate the influence of GSTM1 promoter SNPs on transcriptional activity, eight luciferase reporter constructs were generated. The sequences of these constructs encompassed the three SNPs (ie, −498G>C, −426G>A, and −339C>T) and were transiently transfected into T409 cells. As shown in , reporter gene expression driven by haplotype P−498G-−426G-−339C of GSTM1 was 4.8-fold greater than with haplotype P−498C-−426A-−339T (P < 0.01). There were also statistically significant differences between other haplotypes and haplotype P−498C-−426A-−339T ().
Figure 5 Transient reporter gene expression assays with constructs containing full-length GSTM1 promoter. Luciferase expression of the eight constructs in T409 cells co-transfected with pRL-SV40 to standardize transfection efficiency.
Note: *denotes significantly higher luciferase activity compared to other constructs.
Discussion
This study explored the association between exact copy number of the GSTM1 gene and REC/SPT of HNC in a cohort study and provided compelling evidence of an association between HNC outcome and GSTM1 promoter SNPs/haplotype. One of the major findings is that the exact CNV can predict REC/SPT in a gene-dosage-dependent manner, with subjects having more than two copies of GSTM1 exhibiting the lowest risk of REC/SPT and longest REC/SPT-free survival. Another key finding is that three promoter SNPs (−498C>G, −426A>G, and −339C>T) were significantly associated with REC/SPT in HNC and the haplotype P−498G-−426G-−339C is associated with decreased risk of REC/SPT and improved REC/SPT-free survival. Moreover, it should be noted that all the significant associations remained significant after adjustment for cigarette smoking, alcohol consumption, tumor site, tumor stage, and demographic factors, supporting exact copy number of the GSTM and GSTM1 promoter SNPs/haplotype as independent predictors of REC/SPT in HNC patients.
The incidence of HNC is increasing worldwide, and is associated with high mortality, especially in patients experiencing REC or a SPT.Citation24 Previous studies have suggested that continued smoking, alcohol drinking, and tumor prognostic factors appear to be associated with the likelihood of SPT development.Citation4,Citation25,Citation26 Recently, genetic variants in multiple cellular pathways have been shown to be independent predictors of REC/SPT in HNC patients.Citation20–Citation23 Long-term exposure to certain carcinogens contributes to REC/SPT of HNC, which is an unquestionably smoking-related cancerCitation27–Citation30 and is tightly related to the metabolism of carcinogens. As one of the major hepatic drug-metabolizing enzymes, GSTM1 plays an important role in the detoxification of carcinogens. At time of writing, at least 77 publications have reported on the association between the GSTM1 deletion variant and head and neck squamous cell carcinomas (HNSCC)Citation11,Citation31 and seven publications have examined the association between the GSTM1 deletion variant and outcomes of HNSCC.Citation7,Citation14,Citation32,Citation33 However, these studies have produced conflicting results concerning GSTM1 absence and risk and outcomes of HNSCC.Citation14,Citation30,Citation32,Citation34,Citation35 In our study, we used exact copy number of GSTM1 instead of GSTM1 presence/absence. We found that two or more copies of GSTM1 conferred decreased risk of REC/SPT but one copy did not. In a prostate and bladder cancer study, researchers showed the exact copy number of GSTM1 could predict the risk of bladder cancer, rather than just gene deletion.Citation36 Our finding is also consistent with another study that showed a gene-dosage effect between copy number of GSTM1 and enzyme activity.Citation37
Besides copy number, functional promoter genetic variants could influence gene expression levels by altering the binding ability of transcriptional factors to the gene promoter. In silico prediction modeling indicated multiple putative binding sites of transcriptional factors, such as AP2, GATA1, PEA3, and RXR/RAR, in the promoter region of GSTM1. Moreover, increased active transcriptional activity in the region from −600 to +34 of GSTM1 after response to the Myb gene has been reported.Citation38 In this study, we sequenced the promoter of GSTM1 and found 14 genetic variants, including one insertion variant and 13 SNPs. Furthermore, we demonstrated that three SNPS (−498C>G, −426A>G, and −339C>T) are related to outcomes of HNC. To date, there is one other report related to SNPs in the promoter region of the GSTM1 gene: in that study, Singh and colleagues selected three potentially functional SNPs and found −498C>G was associated with decreased risk of breast cancer.Citation31 This is consistent with the present study in that −498G was related to reduced risk of REC and SPT and was also associated with increased REC/SPT-free survival in HNC. However, Yu et al reported that the −498G allele decreased gene transcription by 30%–40% by reducing the DNA-binding affinity of AP2 for the promoter region.Citation39 This finding is inconsistent with the results of their reported case-control study. Since there are 14 common genetic variants in the promoter region of GSTM1, it is possible that GSTM1 transcription is regulated by multiple transcription factors and could be influenced by several genetic variants in the promoter of GSTM1. In our HNC cohort study, in addition to −498C>G SNP, we also demonstrated that −426A>G and −339T>C were significantly associated with decreased REC/SPT and increased REC/SPT-free survival. This implies the complexity of GSTM1 regulation by different transcriptional factors.
Since three promoter SNPs were independently associated with HNC outcomes, we then constructed haplotypes to examine the concerted effects of these variants. We found significant decreased REC/SPT and increased REC/SPT-free survival in patients with haplotype P−498G-−426G-−339C. Furthermore, in vitro assays demonstrated that haplotype P−498G-−426G-−339C has more transcriptional activity than other haplotypes. These data also support haplotype analysis as a better way of predicting the outcomes of HNC than examining single genetic variants.
Our study has limitations due to its relatively small sample size, so we were not able to assess the combined effects of CNV and promoter haplotypes on HNC outcomes; as such, examination of a larger study population is needed to clarify this relationship. However, this study is ongoing and can therefore be re-examined as more patients are accrued. The findings presented here should also be validated in other populations.
Despite these limitations, our findings support the effect of promoter SNPs and CNV in GSTM1 on the outcomes of HNC. Further study is needed on the regulation of GSTM1 expression and the influence of genetic variants – including CNV, SNP, or insertion – on the expression of GSTM1 in human populations.
Acknowledgements
This work was supported by the Arkansas Biosciences Institute, the University of Arkansas for Medical Sciences’ Clinical Research Center, grant M01 RR14288. Fred Kadlubar passed away on December 4, 2010.
Disclosure
The authors declare no conflicts of interest in this work.
References
- SiegelRNaishadhamDJemalACancer statistics, 2012CA Cancer J Clin2012621102922237781
- LefebvreJLCurrent clinical outcomes demand new treatment options for SCCHNAnn Oncol200516Suppl 6vi7vi1215987995
- ArgirisABrocksteinBEHarafDJCompeting causes of death and second primary tumors in patients with locoregionally advanced head and neck cancer treated with chemoradiotherapyClin Cancer Res20041061956196215041712
- KhuriFRKimESLeeJJThe impact of smoking status, disease stage, and index tumor site on second primary tumor incidence and tumor recurrence in the head and neck retinoid chemoprevention trialCancer Epidemiol Biomarkers Prev200110882382911489748
- SanabriaACarvalhoALVartanianJGMagrinJIkedaMKKowalskiLPFactors that influence treatment decision in older patients with resectable head and neck cancerLaryngoscope2007117583584017473679
- SanabriaACarvalhoALVartanianJGMagrinJIkedaMKKowalskiLPComorbidity is a prognostic factor in elderly patients with head and neck cancerAnn Surg Oncol20071441449145717235712
- HopkinsJCesconDWTseDGenetic polymorphisms and head and neck cancer outcomes: a reviewCancer Epidemiol Biomarkers Prev200817349049918349267
- PearsonWRVorachekWRXuSJIdentification of class-mu glutathione transferase genes GSTM1-GSTM5 on human chromosome 1p13Am J Hum Genet19935312202338317488
- XuSWangYRoeBPearsonWRCharacterization of the human class Mu glutathione S-transferase gene cluster and the GSTM1 deletionJ Biol Chem19982736351735279452477
- RoweJDNievesEListowskyISubunit diversity and tissue distribution of human glutathione S-transferases: interpretations based on electrospray ionization-MS and peptide sequence-specific antiseraBiochem J1997325Pt 24814869230131
- GeislerSAOlshanAFGSTM1, GSTT1, and the risk of squamous cell carcinoma of the head and neck: a mini-HuGE reviewAm J Epidemiol200115429510511447041
- SongDKXingDLZhangLRLiZXLiuJQiaoBPAssociation of NAT2, GSTM1, GSTT1, CYP2A6, and CYP2A13 gene polymorphisms with susceptibility and clinicopathologic characteristics of bladder cancer in Central ChinaCancer Detect Prev2009325–641642319303722
- GolkaKHermesMSelinskiSSusceptibility to urinary bladder cancer: relevance of rs9642880[T], GSTM1 0/0 and occupational exposurePharmacogenet Genomics2009191190390619801959
- OlivieriEHda SilvaSDMendonçaFFCYP1A2*1C, CYP2E1*5B, and GSTM1 polymorphisms are predictors of risk and poor outcome in head and neck squamous cell carcinoma patientsOral Oncol2009459e73e7919442564
- SyamalaVSSreejaLSyamalaVInfluence of germline polymorphisms of GSTT1, GSTM1, and GSTP1 in familial versus sporadic breast cancer susceptibility and survivalFam Cancer20087321322018080216
- PandeySNJainMNigamPChoudhuriGMittalBGenetic polymorphisms in GSTM1, GSTT1, GSTP1, GSTM3 and the susceptibility to gallbladder cancer in North IndiaBiomarkers200611325026116760134
- SharmaAMishraADasBCSardanaSSharmaJKGenetic polymorphism at GSTM1 and GSTT1 gene loci and susceptibility to oral cancerNeoplasma200653430931516830058
- KingRSTeitelCHKadlubarFFIn vitro bioactivation of N-hydroxy-2-amino-alpha-carbolineCarcinogenesis20002171347135410874013
- ColesBFKadlubarFFDetoxification of electrophilic compounds by glutathione S-transferase catalysis: determinants of individual response to chemical carcinogens and chemotherapeutic drugs?Biofactors2003171–411513012897434
- HildebrandtMALippmanSMEtzelCJGenetic variants in the PI3K/PTEN/AKT/mTOR pathway predict head and neck cancer patient second primary tumor/recurrence risk and response to retinoid chemopreventionClin Cancer Res201218133705371322577058
- WangJLippmanSMLeeJJGenetic variations in regulator of G-protein signaling genes as susceptibility loci for second primary tumor/recurrence in head and neck squamous cell carcinomaCarcinogenesis201031101755176120627871
- WuXSpitzMRLeeJJNovel susceptibility loci for second primary tumors/recurrence in head and neck cancer patients: large-scale evaluation of genetic variantsCancer Prev Res (Phila)20092761762419584075
- ZhangXYangHLeeJJMicroRNA-related genetic variations as predictors for risk of second primary tumor and/or recurrence in patients with early-stage head and neck cancerCarcinogenesis201031122118212320819778
- KhuriFRLippmanSMSpitzMRLotanRHongWKMolecular epidemiology and retinoid chemoprevention of head and neck cancerJ Natl Cancer Inst19978931992119017000
- DikshitRPBoffettaPBouchardyCRisk factors for the development of second primary tumors among men after laryngeal and hypopharyngeal carcinomaCancer2005103112326233315852357
- RussoACrosignaniPBerrinoFTobacco smoking, alcohol drinking and dietary factors as determinants of new primaries among male laryngeal cancer patients: a case-cohort studyTumori19968265195259061057
- VineisPPirastuRAromatic amines and cancerCancer Causes Control1997833463559498898
- HashibeMBrennanPStrangeRCMeta- and pooled analyses of GSTM1, GSTT1, GSTP1, and CYP1A1 genotypes and risk of head and neck cancerCancer Epidemiol Biomarkers Prev200312121509151714693745
- ChengLSturgisEMEicherSACharDSpitzMRWeiQGlutathione-S-transferase polymorphisms and risk of squamous-cell carcinoma of the head and neckInt J Cancer199984322022410371337
- MinardCGSpitzMRWuXHongWKEtzelCJEvaluation of glutathione S-transferase polymorphisms and mutagen sensitivity as risk factors for the development of second primary tumors in patients previously diagnosed with early-stage head and neck cancerCancer2006106122636264416703596
- SinghMShahPPSinghAPAssociation of genetic polymorphisms in glutathione S-transferases and susceptibility to head and neck cancerMutat Res20086381–218419418035380
- MatthiasCHarréusUStrangeRInfluential factors on tumor recurrence in head and neck cancer patientsEur Arch Otorhinolaryngol20062631374216003553
- ZafereoMESturgisEMAleemSChaungKWeiQLiGGlutathione S-transferase polymorphisms and risk of second primary malignancy after index squamous cell carcinoma of the head and neckCancer Prev Res (Phila Pa)200925432439
- GeislerSAOlshanAFCaiJWeisslerMSmithJBellDGlutathione S-transferase polymorphisms and survival from head and neck cancerHead Neck200527323224215668931
- WorrallSFCorriganMHighASusceptibility and outcome in oral cancer: preliminary data showing an association with polymorphism in cytochrome P450 CYP2D6Pharmacogenetics1998854334399825835
- NørskovMSFrikke-SchmidtRBojesenSENordestgaardBGLoftSTybjærg-HansenACopy number variation in glutathione-S-transferase T1 and M1 predicts incidence and 5-year survival from prostate and bladder cancer, and incidence of corpus uteri cancer in the general populationPharmacogenomics J201111429229920514077
- McLellanRAOscarsonMAlexandrieAKCharacterization of a human glutathione S-transferase mu cluster containing a duplicated GSTM1 gene that causes ultrarapid enzyme activityMol Pharmacol19975269589659415705
- BartleyPAKeoughRALutwycheJKGondaTJRegulation of the gene encoding glutathione S-transferase M1 (GSTM1) by the Myb oncoproteinOncogene200322487570757514576818
- YuKDDiGHFanLA functional polymorphism in the promoter region of GSTM1 implies a complex role for GSTM1 in breast cancerFASEB J20092372274228719228880