Abstract
Purpose
To investigate the impact of CYP2D6 and CYP2C19 polymorphisms in predicting tamoxifen efficacy and clinical outcomes in Thai breast cancer patients.
Methods
Polymorphisms of CYP2D6 and CYP2C19 were genotyped by the AmpliChip™ CYP450 Test (Roche Molecular Diagnostics, Branchburg, NJ, USA) for 57 patients, who were matched as recurrent versus non-recurrent breast cancers (n = 33 versus n = 24, respectively, with a 5-year follow-up).
Results
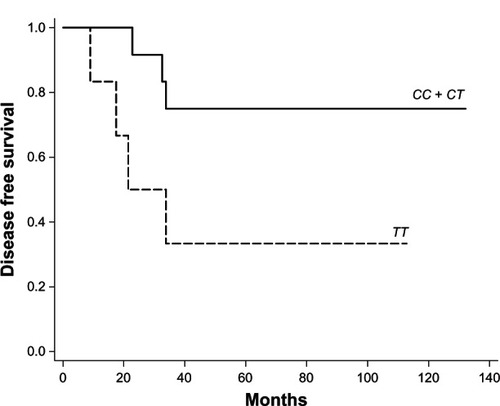
Based on the genotype data, five CYP2D6 predicted phenotype groups were identified in this study including homozygous extensive metabolizer (13 of 57, 22.80%), extensive/intermediate metabolizer (23 of 57, 40.40%), extensive/poor metabolizer (3 of 57, 5.30%), homozygous intermediate metabolizer (14 of 57, 24.50%), and intermediate/poor metabolizer (4 of 57, 7.00%), and three CYP2C19 genotype groups including homozygous extensive metabolizer (27 of 57, 47.40%), extensive/intermediate metabolizer (27 of 57, 47.40%), and homozygous poor metabolizer (3 of 57, 5.30%). The CYP2D6 variant alleles were *10 (52 of 114, 45.60%), *5 (5 of 114, 4.40%), *41 (2 of 114, 1.80%), *4 (1 of 114, 0.90%), and *36 (1 of 114, 0.90%); the CYP2C19 variant alleles were *2 (27 of 114, 23.70%) and *3 (6 of 114, 5.30%). Kaplan–Meier estimates showed significantly shorter disease-free survival in patients with homozygous TT when compared to those with heterozygous CT or homozygous CC at nucleotides 100C>T and 1039C>T (CYP2D6*10) post-menopausal (log-rank test; P = 0.046). They also had increased risk of recurrence, but no statistically significant association was observed (hazard ratio 3.48; 95% confidence interval 0.86–14.07; P = 0.080).
Conclusion
The CYP2D6 and CYP2C19 polymorphisms were not involved in tamoxifen efficacy. However, in the subgroup of post-menopausal women, the polymorphisms in CYP2D6 and CYP2C19 might be useful in predicting tamoxifen efficacy and clinical outcomes in breast cancer patients receiving adjuvant tamoxifen treatment. As the number of breast cancer patients was relatively small in this study, results should be confirmed in a larger group of prospective patients.
Introduction
Tamoxifen is the most commonly prescribed and widely used treatment and adjuvant therapy drug for the prevention of estrogen receptor/progesterone receptor-sensitive breast cancers in pre- and post-menopausal women.Citation1,Citation2 However, approximately 30%–50% of estrogen-positive breast cancer patients have recurrence of the disease and do not respond to tamoxifen treatment.Citation3
Polymorphisms in CYP2D6 and CYP2C19 are clinically important in the metabolism of drugs, as certain allele variants demonstrate either altered activity or nonfunctional enzyme activity with the consequence of 4-hydroxy tamoxifen and endoxifen plasma concentrations.Citation4 Several studies have discovered the association between CYP2D6 and CYP2C19 polymorphisms and plasma concentrations of active metabolites as well as the clinical outcome of breast cancer patients receiving tamoxifen.Citation5,Citation6
It has been reported that European breast cancer patients who receive tamoxifen and are homozygous for CYP2D6*4, thus a poor CYP2D6 metabolizer, have a significantly lower level of endoxifen plasma concentration when compared with homozygous wild type CYP2D6*1.Citation7–Citation9 CYP2D6*10 (100C>T) is the most common intermediate metabolizer allele in the Asian population, which has an allele frequency of approximately 40%–70%. In contrast, Caucasians and African Americans were reported as having approximately a 2%–5% and 3%–8% allele frequency, respectively.Citation10–Citation12
The CYP2D6*10 homozygous variant genotype could affect the efficacy of tamoxifen, and it is associated with significantly lower plasma concentrations of 4-hydroxy tamoxifen when compared with the homozygous wild type genotype. Also, it was found that breast cancer patients with the CYP2D6*10 homozygous variant genotype had a significantly worse disease-free survival (DFS) than those with heterozygous (CT) or homozygous wild type genotype.Citation13–Citation15 Lim et al performed modeling analysis to investigate the influence of CYP2D6, genotype CYP3A5, CYP2C9, and CYP2C19 polymorphisms on tamoxifen pharmacokinetics and found that CYP2D6*5/*10 and *10/*10 were significantly associated with lower concentrations of endoxifen and N-desmethyl tamoxifen.Citation16 The CYP2C19 gene has two major poor metabolizer (PM) alleles that result in deficiency of the enzyme. However, information is limited on the possibility of the CYP2C19 genotype affecting the efficacy of tamoxifen, but the result from van Schaik et al demonstrated that CYP2C19 is associated with increased survival in breast cancer patients using tamoxifen.Citation17
Therefore, this study aimed to identify the polymorphisms in CYP2D6 and CYP2C19 in patients with breast cancer and to investigate the impact of genetic polymorphisms on disease recurrence in patients who received adjuvant tamoxifen.
Material and methods
Clinical subjects
Fifty-seven participants in this retrospective study were recruited from a primary recurrent and non-recurrent breast cancer population enrolled between February 1997 and January 2008 at the Department of Medicine, Ramathibodi Hospital in Bangkok, Thailand. All 57 patients were assigned randomly to receive 20 mg/day adjuvant tamoxifen for 5 years. This study was designed for 33 breast cancer recurrence and 24 breast cancer non-recurrence. The two groups were matched by the characteristics of the patients (). Patients receiving selective serotonin reuptake inhibitors were excluded in the post hoc analyses. Written informed consent forms were obtained from all patients. The study was approved by the Ramathibodi Hospital Ethics Committee.
Table 1 Characteristics of non-recurrent and recurrent breast cancer patients
Patient characteristics
The use of adjuvant tamoxifen was similar in the two groups (cases and controls) (). The mean age of the subjects was 48.9 ± 10.6 years. The median follow-up time of the case and control group was 93.5 months (range 59.0–172.0) and 22.0 months (range 2.0–62.0), respectively. The median follow-up time was 48.0 months (range 2.0–172.0). The number of pre- and post-menopausal patients was 38 and 19, respectively. All patients were estrogen receptor-positive except for one patient, who was estrogen receptor-negative but progesterone receptor-positive. Among the 33 patients with breast cancer recurrence, 6.06% (2/33) were human epidermal growth factor receptor-2 (Her-2)-positive and 60.60% (20/33) were of unknown status. Twenty-five (43.80%; 25/57) patients had positive axillary lymph nodes. Most patients were treated with a modified radical mastectomy. The adjuvant chemotherapy comprised cyclophosphamide, intravenous methotrexate, and 5-fluorouracil, and Adriamycin®-based and Adriamycin–taxane-based regimens. Three patients in this study did not receive adjuvant chemotherapy, despite their eligibility for treatment, because they had positive lymph node (N1) axillaries (two patients in the control arm and one patient in the case arm, respectively). There was no significant difference in patient characteristics between non-recurrent and recurrent breast cancers ().
Analysis of polymorphisms in CYP2D6 and CYP2C19
Genomic DNA was extracted from ethylenediaminetetraacetic acid blood and isolated by the salting out procedure.Citation17 The microarray technique (AmpliChip™ CYP450 Test; Roche Molecular Diagnostics, Branchburg, NJ, USA) was used for detection of polymorphisms in CYP2D6 and CYP2C19 according to the manufacturer’s instructions. The main process of the test comprised polymerase chain reaction amplification, fragmentation and labeling, hybridization, staining, and scanning. The test explored 29 known polymorphisms in the CYP2D6 gene, including gene deletion and duplication, and 33 different alleles were acceptable for identification. The CYP2D6 genotypes were classified based on previous studies.Citation18–Citation20 There were four phenotypic categories according to allele-related enzyme activity: no enzyme activity alleles (PM) *3, *4, *5, *6, *7, *8, *11, *14A, *15, *19, *20, *36, *40, and *4XN; decreased enzyme activity alleles (intermediate metabolizer) *9, *10, *17, *29, *41,*10XN, *17XN, and *41XN; normal enzyme activity alleles (extensive metabolizer) *1, *2, and *35; and increased enzyme activity alleles (ultra-rapid metabolizer) *1XN, *2XN, and *35XN. The polymorphisms in CYP2C19 were genotyped for *1, *2, and *3.
Statistical analysis
Descriptive statistics were used to describe the clinical characteristics of the subjects. Hardy–Weinberg equilibrium was conducted with Haploview 4.2 (Broad Institute of Harvard and MIT, Cambridge, MA, USA). Fisher’s exact test or Pearson’s Chi-squared test was used to compare the different alleles and patient characteristics between recurrent and non-recurrent breast cancers. DFS was defined as the time from surgery to the recurrence of breast cancer event (local, regional, or distant occurrence or contralateral breast cancer) or death from any cause. Patients who were alive without a breast cancer relapse were censored at the last follow-up date. Survival curves were estimated with the Kaplan–Meier method. Statistical significance of a relationship between breast cancer outcomes and each of the genetic polymorphisms was compared by the log-rank test. The univariate Cox proportion hazard model was used to estimate the hazard ratio (HR) for comparing the genotype of each group. All tests were two-sided and P-values of less than 0.05 were considered statistically significant. Statistical analyses were conducted using Stata® version 12 (StataCorp L P, College Station, TX, USA).
Results
Allele frequencies of the CYP2D6 and CYP2C19
The polymorphisms observed in CYP2D6 and CYP2C19 were in Hardy–Weinberg equilibrium and they matched those in a previous report on Asian populations. shows the frequencies of CYP2D6 alleles among different ethnic groups. The CYP2D6*10 and CYP2D6*5 (gene deletion) alleles were the most variant and nonfunctional, respectively, in this study, with variance and allele frequency of 45.6% and 4.40%, respectively. Rare variant alleles found that CYP2D6*36 and *41 had a frequency of 0.90% and 1.80%, respectively. The results showed that the CYP2D6*4 allele with a frequency of 0.90% was characterized by a 1846G>A mutation. The frequencies of CYP2C19 alleles are shown in . The CYP2C19*2 allele was the most common variant found in this study at 23.70%. There were no significant differences in allelic frequencies of CYP2D6 and CYP2C19 between recurrent and non-recurrent breast cancers (Table S1).
Table 2 Frequencies of the CYP2D6Citation10,Citation11,Citation24,Citation25 and CYP2C19Citation26 allele in different ethnic groups
Frequencies of the genotype and predicted phenotype of CYP2D6 and CYP2C19
Most of the CYP2D6 genotypes presented with heterozygous and homozygous intermediate metabolizer alleles. For example, CYP2D6*1/*10 and *10/*10 had allele frequencies of 28.10% (16/57) and 22.80% (13/57), respectively. Allele frequencies of the CYP2D6 genotypes were 15.70% for CY2D6*1/*1 (9/57), 3.50% for *1/*2 (2/57), 3.50% for *1/*5 (2/57), 1.80% for *1/*36 (1/57), 1.80% for *1/*41 (1/57), 3.50% for *2/*2 (2/57), 1.80% for *2/*4 (1/57), 7.00% for *2/*10 (4/57), 5.20% for *10/*5 (3/57), 1.80% for *10/*14B (1/57), 1.80% for *10/*35 (1/57), and 1.80% for *10/*41 (1/57) (Table S2). Additionally, no homozygous PM or multiple copy (ultra-rapid metabolizer) of CYP2D6 alleles were observed in this study ().
Table 3 CYP2D6 and CYP2C19 predicted phenotype according to non-recurrence and recurrence groups
Frequency of the homozygous CYP2C19*1 and homozygous PM allele of the CYP2C19 genotype was 47.40% and 5.30% for *1/*1 (27/57) and *2/*2 (3/57), respectively. Frequency of the remaining CYP2C19 genotypes was 36.80% and 10.50% for *1/*2 (21/57) and *1/*3 (6/57), respectively (Table S2). In addition, , S2, and S3 shows no significant difference in the distribution of CYP2D6 and CYP2C19 genotypes and predicted phenotypes between recurrent and non-recurrent breast cancers.
CYP2D6 and CYP2C19 polymorphisms and breast cancer recurrence
The time it took for the patients to develop breast cancer recurrence was evaluated using Kaplan–Meier analysis. Kaplan–Meier estimates showed significantly shorter DFS () in patients with homozygous TT when compared to those with heterozygous CT or homozygous CC at nucleotides 100C>T and 1039C>T (CYP2D6*10) in post-menopausal women (log-rank test; P = 0.046 and P = 0.046), in which two single nucleotide polymorphisms were in linkage disequilibrium. In addition, patients with CYP2D6*10/*10 followed a different trend for DFS when compared to heterozygous CYP2D6*10 and homozygous wild type (CYP2D6 Wt/Wt) in post-menopausal women, but there was no statistical significance (P = 0.087). Finally, no statistically significant difference in DFS was detectable in other nucleotides or genotypes of CYP2D6 and CYP2C19 (Tables S4 and S5).
Risk estimation between genotypes of CYP2D6 and CYP2C19
Patients with heterozygous GA at nucleotide 1846G>A (CYP2D6*4) showed an increased risk of recurrence, but no overall statistically significant difference was observed in pre-menopausal patients (HR 5.82; 95% confidence interval [CI] 0.74–46.02; P = 0.094 and HR 5.84; 95% CI 0.70–48.55; P = 0.102). Overall, post-menopausal patients with homozygous TT at nucleotide 100C>T and 1039C>T (CYP2D6*10) tended to have increased risk of recurrence, but no statistically significant association was observed. In contrast, pre-menopausal patients with homozygous TT at nucleotides 100C>T and 1039C>T tended to have decreased risk of recurrence, but no significant association was observed (Table S6). On the other hand, the results showed that pre-menopausal patients with heterozygous GC at nucleotide 4180G>C had decreased risk of developing recurrence when compared to patients with homozygous GG (HR 0.48; 95% CI 0.20–1.15; P = 0.099). shows that the genotype of CYP2D6 and CYP2C19 had increased risk of developing recurrence, but no statistically significant association was observed.
Table 4 Risk estimation between CYP2D6 and CYP2C19 genotypes and recurrences in breast cancer patients among overall, pre-menopausal, and post-menopausal groups
Discussion
This study aimed to investigate the association between CYP2D6 and CYP2C19 polymorphisms and breast cancer outcomes in Thai female breast cancer patients treated with tamoxifen. The characteristics of breast cancer patients may affect the clinical outcome.
Overall, the presence of variant CYP2D6 and CYP2C19 alleles had no significant difference in DFS between recurrent and non-recurrent breast cancers. However, Kaplan–Meier estimates showed a significant difference in DFS in patients with homozygous variant (TT) when compared with heterozygous variant (CT) or homozygous wild type (CC) at nucleotides 100C>T and 1039C>T (CYP2D6*10) in post-menopausal patients (log-rank test P = 0.046 and P = 0.046), in which two single nucleotide polymorphisms were associated with linkage disequilibrium.
Previous studies investigated the association between polymorphisms in CYP2D6 and tamoxifen efficacy and clinical outcomes in patients receiving adjuvant tamoxifen.Citation14,Citation15 Goetz et al initially reported that breast cancer patients with decreased CYP2D6 metabolism had a significantly shorter recurrence time (HR 1.91; 95% CI 1.05–3.45; P = 0.034) and worse relapse-free survival (HR 1.74; 95% CI 1.10–2.74; P = 0.017) when compared to patients with extensive CYP2D6 metabolism. Patients with the PM phenotype (CYP2D6*4/*4) had a significantly higher risk of breast cancer relapse approximately three times that of patients with extensive metabolizers (CYP2D6*1/*1 and *1/*4) (HR 3.12; P = 0.007).Citation22 Xu et al showed that patients with the CYP2D6*10 homozygous TT genotype had significantly worse DFS than those with the heterozygous CT and homozygous CC genotype (HR 4.7; 95% CI 1.1–20.0; P = 0.004).Citation13 Lim et al reported that patients with the CYP2D6*10/*10 genotype had a significantly higher risk of breast cancer relapse within 10 years after surgery when compared to those with other genotypes (time to progression 5.03 versus 21.8 months, P = 0.0032).Citation23 Kiyotani et al reported that patients with CYP2D6*10/*10 and CYP2D6*1/*10 showed significantly shorter recurrence-free survival when compared to those with CYP2D6*1/*1 (HR 9.52; 95% CI 2.79–32.45; P = 0.000036).Citation24
In contrast, previous studies from both European and Asian populations showed no significant association between polymorphisms in CYP2D6 and outcome of tamoxifen treatment. In the first, Okishiro et al reported no significantly different relapse-free survival rates between breast cancer patients with CYP2D6*10/*10 genotypes and those with CYP2D6*1/*1 or CYP2D6*1/*10 genotypes, nor was there a difference between patients with CYP2C19 PM genotypes (CYP2C19*2/*2, *2/*3, or *3/*3) and those with CYP2C19 extensive metabolizer genotypes (CYP2C19 *1/*1, *1/*2, or *1/*3).Citation25 Toyama et al demonstrated no significant correlation between patients with the CYP2D6*10/*10 genotype and survival time (DFS, distant DFS, breast cancer-specific survival, and overall survival) when compared to those with CYP2D6 *1/*1 and *1/*10 genotypes.Citation26 In contrast, a report from Sweden showed that patients with the CYP2D6*4/*4 genotype had significantly better DFS than those with heterozygous or homozygous CYP2D6*1 (P = 0.004 and P = 0.005, respectively).Citation27
The data in this study support the conclusion that CYP2D6 and CYP2C19 variants are not significantly associated with the clinical outcome in breast cancer patients taking adjuvant tamoxifen. Conversely, in a group of post-menopausal women, the polymorphisms in CYP2D6*10/*10 might be useful in predicting tamoxifen efficacy and clinical outcomes when compared to heterozygous CYP2D6*10 and homozygous wild type (CYP2D6 *1/*1).
However, this study had some limitations. Primarily, the retrospective nature of the study design is weak, which it shares with all other available studies. This retrospective method also lacks data correlation between polymorphisms in CYP2D6 and the plasma concentration of tamoxifen metabolites. While the small sample size and low number of PM phenotypes in this study may have given a low statistical power, all samples collected from the recruited were matched in a case–control manner.
It is possible that one or more of these variants are associated with a specific subgroup of breast cancer patients. The data in this study showed that the high frequency of CYP2D6*10 is similar to Asian populations reported previously,Citation9,Citation21 and only nine variations include gene deletion, gene conversion, 1584C>G, 100C>T, 1039C>T, 1661G>C, 1846G>A, 2850C>T, and 4180G>C. No homozygous CYP2D6 PM (CYP2D6*3, *4, and *5) or homozygous ultra-rapid metabolizers (CYP2D6*1XN, *2XN, and *35XN) in this study is due possibly to the small sample size.
Conclusion
The variant alleles of CYP2D6 and CYP2C19 genes in this study were not involved in tamoxifen efficacy. However, in the subgroup of post-menopausal women, the polymorphisms in CYP2D6 and CYP2C19 might be useful in predicting tamoxifen efficacy and clinical outcomes in breast cancer patients receiving adjuvant tamoxifen treatment. As the number of breast cancer patients was small in this study, results should be confirmed in a larger group of patients.
Acknowledgments
This study was supported by a grant from the Thailand Center of Excellent Life Science (TCELS). The authors would like to thank Dr Suwit J Somponpun for assistance in the preparation and editing of this manuscript. The authors are also grateful to all participants who contributed to the study.
Supplementary table
Table S1 CYP2D6 and CYP2C19 alleles frequency compared between groups
Table S2 CYP2D6 and CYP2C19 genotypes frequency compared between groups
Table S3 Genotype frequencies of CYP2D6 and CYP2C19 of 33 breast cancer recurrence and 24 breast cancer non-recurrence cases
Table S4 Log-rank test of CYP2D6 genotypes
Table S5 Log-rank test of CYP2C19 genotypes
Table S6 Risk estimation between genotypes and recurrences in breast cancer patients
Disclosure
The authors report no conflicts of interest in this work.
References
- FisherBCostantinoJPWickerhamDLTamoxifen for the prevention of breast cancer: current status of the National Surgical Adjuvant Breast and Bowel Project P-1 studyJ Natl Cancer Inst200597221652166216288118
- ColleoniMGelberSGoldhirschATamoxifen after adjuvant chemotherapy for premenopausal women with lymph node-positive breast cancer: International Breast Cancer Study Group Trial 13-93J Clin Oncol20062491332134116505417
- OsborneCKTamoxifen in the treatment of breast cancerN Engl J Med199833922160916189828250
- DalyAKPharmacogenetics of the major polymorphic metabolizing enzymesFundam Clin Pharmacol2003171274112588628
- Ingelman-SundbergMSimSCGomezARodriguez-AntonaCInfluence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspectsPharmacol Ther2007116349652618001838
- HoskinsJMCareyLAMcLeodHLCYP2D6 and tamoxifen: DNA matters in breast cancerNat Rev Cancer20099857658619629072
- StearnsVJohnsonMDRaeJMActive tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetineJ Natl Cancer Inst200395231758176414652237
- JinYDestaZStearnsVCYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatmentJ Natl Cancer Inst2005971303915632378
- LimYCLiLDestaZEndoxifen, a secondary metabolite of tamoxifen, and 4-OH-tamoxifen induce similar changes in global gene expression patterns in MCF-7 breast cancer cellsJ Pharmacol Exp Ther2006318250351216690721
- KubotaTYamauraYOhkawaNHaraHChibaKFrequencies of CYP2D6 mutant alleles in a normal Japanese population and metabolic activity of dextromethorphan O-demethylation in different CYP2D6 genotypesBr J Clin Pharmacol2000501313410886115
- SachseCBrockmollerJBauerSRootsICytochrome P450 2D6 variants in a Caucasian population: allele frequencies and phenotypic consequencesAm J Hum Genet19976022842959012401
- GrieseEUAsante-PokuSOfori-AdjeiDMikusGEichelbaumMAnalysis of the CYP2D6 gene mutations and their consequences for enzyme function in a West African populationPharmacogenetics19999671572310634134
- XuYSunYYaoLAssociation between CYP2D6 *10 genotype and survival of breast cancer patients receiving tamoxifen treatmentAnn Oncol20081981423142918407954
- SirachainanEJaruhathaiSTrachuNCYP2D6 polymorphisms influence the efficacy of adjuvant tamoxifen in Thai breast cancer patientsPharmgenomics Pers Med2012514915323226070
- SukasemCSirachainanEChamnanphonMImpact of CYP2D6 polymorphisms on tamoxifen responses of women with breast cancer: a microarray-based study in ThailandAsian Pac J Cancer Prev20121394549455323167378
- LimJSChenXASinghOImpact of CYP2D6, CYP3A5, CYP2C9 and CYP2C19 polymorphisms on tamoxifen pharmacokinetics in Asian breast cancer patientsBr J Clin Pharmacol201171573775021480951
- van SchaikRHKokMSweepFCThe CYP2C19*2 genotype predicts tamoxifen treatment outcome in advanced breast cancer patientsPharmacogenomics20111281137114621830868
- MillerSADykesDDPoleskyHFA simple salting out procedure for extracting DNA from human nucleated cellsNucleic Acids Res198816312153344216
- SchrothWHamannUFaschingPACYP2D6 polymorphisms as predictors of outcome in breast cancer patients treated with tamoxifen: expanded polymorphism coverage improves risk stratificationClin Cancer Res201016174468447720515869
- MurdterTESchrothWBacchus-GerybadzeLActivity levels of tamoxifen metabolites at the estrogen receptor and the impact of genetic polymorphisms of phase I and II enzymes on their concentration levels in plasmaClin Pharmacol Ther201189570871721451508
- GoetzMPKnoxSKSumanVJThe impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifenBreast Cancer Res Treat2007101111312117115111
- LimHSJu LeeHSeok LeeKSook LeeEJangIJRoJClinical implications of CYP2D6 genotypes predictive of tamoxifen pharmacokinetics in metastatic breast cancerJ Clin Oncol200725253837384517761971
- KiyotaniKMushirodaTSasaMImpact of CYP2D6*10 on recurrence-free survival in breast cancer patients receiving adjuvant tamoxifen therapyCancer Sci200899599599918294285
- OkishiroMTaguchiTJin KimSShimazuKTamakiYNoguchiSGenetic polymorphisms of CYP2D6*10 and CYP2C19*2,*3 are not associated with prognosis, endometrial thickness, or bone mineral density in Japanese breast cancer patients treated with adjuvant tamoxifenCancer2009115595296119156902
- ToyamaTYamashitaHSugiuraHKondoNIwaseHFujiiYNo association between CYP2D6*10 genotype and survival of node-negative Japanese breast cancer patients receiving adjuvant tamoxifen treatmentJpn J Clin Oncol2009391065165619596663
- WegmanPElingaramiSCarstensenJStalONordenskjoldBWingrenSGenetic variants of CYP3A5, CYP2D6, SULT1A1, UGT2B15 and tamoxifen response in postmenopausal patients with breast cancerBreast Cancer Res200791R717244352
- ZangerUMRaimundoSEichelbaumMCytochrome P450 2D6: overview and update on pharmacology, genetics, biochemistryNaunyn Schmiedebergs Arch Pharmacol20043691233714618296