Abstract
Single-nucleotide polymorphisms (SNPs) in the gene coding for the efflux-transport protein ABCB1 (P-glycoprotein) are commonly inherited as haplotypes. ABCB1 SNPs and haplotypes have been suggested to influence the pharmacokinetics and therapeutic outcome of the tyrosine kinase inhibitor (TKI) imatinib, used for treatment of chronic myeloid leukemia (CML). However, no consensus has yet been reached with respect to the significance of variant ABCB1 in CML treatment. Functional studies of variant ABCB1 transport of imatinib as well as other TKIs might aid the interpretation of results from in vivo association studies, but are currently lacking. The aim of this study was to investigate the consequences of ABCB1 variant haplotypes for transport and efficacy of TKIs (imatinib, its major metabolite N-desmethyl imatinib [CGP74588], dasatinib, nilotinib, and bosutinib) in CML cells. Variant haplotypes – including the 61A>G, 1199G>A, 1236C>T, 1795G>A, 2677G>T/A, and 3435T>C SNPs – were constructed in ABCB1 complementary DNA and transduced to K562 cells using retroviral gene transfer. The ability of variant cells to express ABCB1 protein and protect against TKI cytotoxicity was investigated. It was found that dasatinib and the imatinib metabolite CGP74588 are effectively transported by ABCB1, while imatinib, nilotinib, and bosutinib are comparatively weaker ABCB1 substrates. None of the investigated haplotypes altered the protective effect of ABCB1 expression against TKI cytotoxicity. These findings imply that the ABCB1 haplotypes investigated here are not likely to influence TKI pharmacokinetics or therapeutic efficacy in vivo.
Introduction
The tyrosine kinase inhibitors (TKIs) imatinib, dasatinib, and nilotinib are indicated for first-line treatment of chronic myeloid leukemia (CML). Moreover, dasatinib and nilotinib together with bosutinib are indicated for the treatment of patients with resistance or intolerance to first-line therapy. In general, most patients respond well to therapy, although a significant proportion of CML patients have to dose-adjust or switch therapies due to adverse events or suboptimal response.Citation1,Citation2 In order to ensure a more personalized CML treatment strategy, a deeper understanding of the factors that determine response and resistance to the individual TKIs is required.
Imatinib, dasatinib, and nilotinib have previously been reported as substrates for the efflux-transport protein ABCB1 (P-glycoprotein).Citation3–Citation5 Less is known about bosutinib efflux, although a single report indicates that bosutinib is unlikely to be transported by ABCB1.Citation4 In addition, the major imatinib metabolite N-desmethyl imatinib (CGP74588) was also shown to be an ABCB1 substrate.Citation6 This metabolite is pharmacologically active in vitro, although less potent than imatinib.Citation6–Citation8
ABCB1 is expressed in tissues that are involved in the absorption and elimination of TKIs, including the intestine, liver, and kidneys.Citation9 ABCB1 expression has also been found in CML stem cells and in the circulating leukocytes of CML patients.Citation10–Citation12 Consequently, in addition to influencing the influx–efflux of active drug into target cells, ABCB1-transport activity might influence the systemically circulating TKI concentration that reaches target cells. The influx-transport activity of the organic cation transporter 1 (OCT-1) as well as imatinib plasma concentration have previously been correlated with the outcome of CML therapy.Citation13–Citation15 These findings indicate that analysis of drug-transport activity, or markers thereof, might be useful predictors of response to imatinib, and perhaps to the second-generation TKIs as well.
The ABCB1 gene is highly polymorphic, with about 100 identified single-nucleotide polymorphisms (SNPs) located in the coding regions, some of which have been associated with the efflux, pharmacokinetics, or therapeutic outcome of several drug classes.Citation16 Moreover, at least 28 coding and noncoding ABCB1 SNPs are frequently inherited together, defining distinct haplotypes.Citation17 The most commonly studied haplotype consists of the 1236C>T, 2677G>T/A, and 3435T>C SNPs. These SNPs have been evaluated for their influence on imatinib plasma concentrations and therapeutic efficacy in CML patients. However, the reports are inconclusive: some show influence of the individual SNPs or the complete haplotype on plasma concentrations and/or therapeutic outcome of imatinib,Citation18–Citation21 while others do not.Citation22–Citation25 In addition, there are other nonsynonymous ABCB1 SNPs that have been associated with the therapeutic outcome of ABCB1 substrate drugs,Citation26,Citation27 but that have not yet been studied with regard to TKI transport. In light of contradictory results and the ongoing debate about ABCB1 SNPs and their significance in CML treatment, a functional study of ABCB1 SNPs in relation to their influence on TKI transport is needed. Therefore, the aim of this study was to investigate the influence of ABCB1 variant haplotypes on TKI transport and efficacy. For that purpose, the ABCB1 SNPs 61A>G, 1199G>A, 1236C>T, 1795G>A, 2677G>T/A, and 3435T>C were constructed in combinations to result in variant haplotypes that were transduced to a CML cell line. The impact of ABCB1 variant haplotypes on transport and efficacy of imatinib, CGP74588, dasatinib, nilotinib, and bosutinib was investigated.
Materials and methods
Drugs and chemicals
Imatinib and CGP74588 were provided by Novartis Pharma (Basel, Switzerland). Dasatinib, nilotinib, and bosutinib were purchased from Selleck Chemicals (Houston, TX, USA). Stock solutions of 10 mM were prepared for all drugs, stored at −20°C, aliquoted to avoid repeated freeze–thawing, and were used within 1 year of preparation. Imatinib and CGP74588 stock solutions were prepared in water, while nilotinib, dasatinib, and bosutinib were prepared in dimethyl sulfoxide. Unless otherwise stated, all chemicals used in this study were purchased from Sigma-Aldrich (St Louis, MO, USA).
Cells and culturing conditions
The CML cell line K562 (LGC Standards, Teddington, UK) was used for ABCB1 transduction and parental as well as transduced cell lines were kept in Roswell Park Memorial Institute 1640 medium supplemented with penicillin, streptomycin, and 10% fetal bovine serum (FBS). Human embryonic kidney 293T cells (LGC Standards) were cultured in Dulbecco’s Modified Eagle’s Medium, supplemented with penicillin, streptomycin, and 10% heat-inactivated FBS. Cell-culture reagents were purchased from Life Technologies, Paisley, UK. All cell lines were verified to be mycoplasma-free.
ABCB1 single-nucleotide polymorphisms and haplotypes
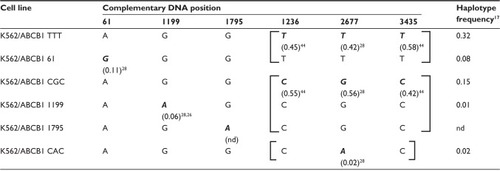
The ABCB1 SNPs studied here were selected based on their single-nucleotide substitution, location in the coding region of the ABCB1 gene, and a minor allele frequency of >2% in the Caucasian population. SNP and haplotype frequencies were obtained from previously published population studiesCitation17,Citation26,Citation28 or from the National Center for Biotechnology Information dbSNP database. The ABCB1 SNPs included in this study were 61A>G (rs9282564), 1199G>A (rs2229109), 1236C>T (rs1128503), 1795G>A (rs2235036), 2677G>T/A (rs2032582), and 3435T>C (rs1045642). The 1236C>T, 2677G>T/A, and 3435T>C SNPs are in linkage disequilibrium and are commonly inherited together as one of the two haplotypes (1236T, 2677T, 3435T) or (1236C, 2677G, 3435C), referred to here as the TTT or CGC haplotype. In approximately 2% of the Caucasian population, 2677G>T is substituted by an A, giving rise to the CAC haplotype (1236C, 2677A, 3435C).Citation17,Citation26 The 1236 and 3435 SNPs are silent substitutions; given the method of retroviral gene transfer with artificial transcriptional regulation, our main focus was to study the posttranslational effects of ABCB1 variants. However, the complete haplotypes of 1236, 2677, and 3435 SNPs were constructed to ensure that any differences between the variant cell lines had not been caused by altered efficacy of translation due to linked silent SNPs in the ABCB1 transcript. Consequently, the three haplotypes were constructed in separate vectors for transduction into K562 cells. The 61A>G and 1199G>A SNPs were constructed together with TTT and CGC, respectively, the haplotypes with which they are most frequently associated.Citation17 The 1795G>A SNP was included in the study because it was already in its variant form in the transcript used for vector constructs. To the best of our knowledge, this SNP has not been associated with a specific haplotype, thus it was kept together with the CGC haplotype, since this was the haplotype of the transcript used for vector constructs. An overview of the genotypes of generated cell lines and frequencies of minor alleles and haplotypes appears in .
Table 1 ABCB1 genotype of transduced cell lines
ABCB1 complementary DNA (cDNA; GenBank ID NM_000927.3) in a pCMV6-XL4 vector (OriGene, Rockville, MD, USA) was used for incorporation of variant nucleotides corresponding to each specific SNP, using the QuickChange II Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA). Incorporation of variants and ABCB1 reference sequence was confirmed by automated Sanger sequencing, using the services of GATC Biotech (Konstanz, Germany).
Generation of K562 cells with variant ABCB1 expression
ABCB1 cDNA was ligated into a murine stem cell virus-IRES-enhanced yellow fluorescent protein (EYFP) retroviral vector (MIY),Citation29 using the Rapid DNA Dephos and Ligation Kit (Roche, Basel, Switzerland). The correct orientation of the gene insert was confirmed by Sanger sequencing (GATC Biotech). The MIY-ABCB1 vector (11 kb long) exceeded the optimal size of insert cDNA to the retroviral transfer system and had to be shortened for efficient transductions. The restriction enzyme PmeI (New England Biolabs, Ipswich, MA, USA) was used to cleave off a 750-base-long fragment in the vector–gene complex from position +199 in the ABCB1 3′-untranslated region into nonsignificant parts of the vector. MIY-ABCB1 (4 μg) and 2 μg each of the helper vectors vesicular stomatitis virus glycoprotein G and Pol-Gag were mixed with a final concentration of 125 mM CaCl2, followed by calcium phosphate transfection of 1 × 10Citation6 293T cells. Viral 293T supernatants were collected over 48 hours and filtered through a 0.45 μm sterile cellulose acetate filter (Whatman, Dassel, Germany). K562 cells (0.5 × 10Citation6) were transduced with viral supernatants using spin infection (1.5 hours in 1200 g, 22°C) in the presence of 4 μg/mL polybrene. EYFP+/ABCB1+ cells were sorted using flow cytometry. Sorting aimed at ensuring equal median fluorescence intensities (MFIs) of EYFP between cell lines. An empty MIY vector was transduced to K562 cells to obtain the control cell line, referred to here as K562/ve.
Cell-membrane expression of ABCB1 protein
The expression level of ABCB1 protein in the cell membrane after retroviral gene transfer was evaluated using flow cytometry. Cells (1 × 10Citation6) were labeled with 0.25 μg of primary mouse antihuman ABCB1, clone 17F9 (BD Biosciences, San Jose, CA, USA), washed twice in phosphate-buffered saline supplemented with 2% FBS, followed by labeling with 0.5 μg of secondary goat antimouse immunoglobulin G2b conjugated with allophycocyanin/Cy7 (Abcam, Cambridge, UK). Cells were washed once more in phosphate-buffered saline before simultaneous detection of ABCB1 and EYFP on a Gallios flow cytometer (Beckman Coulter, Bromma, Sweden).
Efficacy of TKIs and vincristine in variant ABCB1 cells
All cell lines were investigated in terms of resistance to TKIs, using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay,Citation30 performed as described in a previous report.Citation8 In addition, the resistance of K562, K562/ve, and K562/ABCB1 TTT to vincristine was investigated in order to verify the functionality of transduced ABCB1. Vincristine is a vinca alkaloid that is commonly used in chemotherapy and is a known ABCB1 substrate.Citation16,Citation31 Experiments were performed in a total of nine replicates for TKIs and six replicates for vincristine. A dose-response regression with variable slope and a top plateau constrained to ,100% was fitted to each of the replicates in GraphPad Prism 5 (GraphPad Software, La Jolla, CA, USA). Halfmaximal inhibitory concentration (IC50) values were extracted from each dose-response regression; the mean IC50 and 95% confidence intervals of the same were calculated and used for comparisons of drug sensitivity between cell lines with different ABCB1 variant haplotypes. Of note, the MTT assay reflects the number of living cells rather than dead cells, and thus the obtained IC50 values reflect the sum of cell death and inhibition of proliferation caused by TKIs.
Intracellular accumulation of TKIs in ABCB1-expressing cells
In order to confirm that ABCB1 drug efflux was involved in the TKI resistance observed using the MTT assay, parental K562 cells, as well as K562/ve and K562/ABCB1 TTT, were incubated with TKIs until influx–efflux equilibrium was attained. Intracellular accumulation of TKIs was quantified using an ultraperformance liquid chromatography (UPLC) tandem mass-spectrometry method (see Supplementary materials for details).
Statistical analysis
Differences in ABCB1 protein expression and intracellular TKI accumulation between cell lines were analyzed with Student’s independent t-tests. P-values <0.05 were considered significant.
Results
Characterization of ABCB1 transduced cell lines
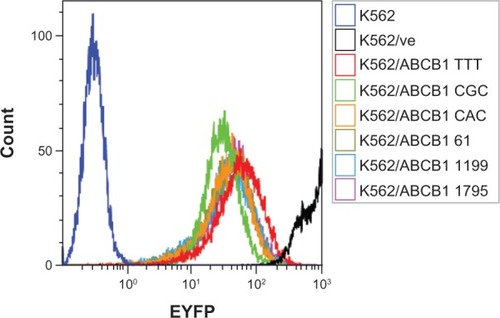
In this study, we used the MIY retroviral vector containing an internal ribosome entry site between the ABCB1 cDNA and the reporter gene EYFP to ensure the same protein-translation level of both genes in transduced cells. K562 cells transduced with variants of ABCB1 were sorted based on equal EYFP protein expression. Flow-cytometry analysis showed that similar EYFP fluorescence profiles had been obtained in all ABCB1 transduced cell lines (). The EYFP MFI ranged from 24.8 for K562/ABCB1 CGC to 55.3 for K562/ABCB1 TTT. K562/ve had an extremely high EYFP expression (MFI = 1,017), which may be related to more efficient translation of this shorter transcript compared to EYFP–ABCB1 transcripts.
Figure 1 Expression of enhanced yellow fluorescent protein (EYFP). All ABCB1 transduced cell lines had similar expression of EYFP, as shown by the overlapping fluorescent profiles. Cells transduced with empty vector (K562/ve) express high EYFP fluorescence.
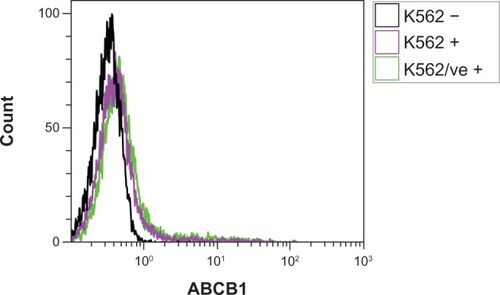
ABCB1 MFI, normalized to EYFP MFI, revealed that none of the variant cell lines, with the exception of K562/ABCB1 CGC, had significantly different quantities of ABCB1 expressed in the membrane than K562/ABCB1 TTT (). However, upon analyzing the uncorrected ABCB1 MFI, all variant cell lines had significantly lower ABCB1 membrane expression than K562/ABCB1 TTT (). Parental K562 cells and K562/ve did not express ABCB1 in the cell membrane, as demonstrated by the fact that these cell lines had similar mean ABCB1 MFI (MFIs = 0.45 and 0.52) as unlabeled K562 (MFI = 0.35) ().
Figure 2 (A and B) Expression of ABCB1 in cell membranes. Parental cells (K562), K562 transduced with empty vector (K562/ve), and ABCB1 variant haplotypes were analyzed in parallel for expression of ABCB1 and the reporter protein, enhanced yellow fluorescent protein (EYFP). (A) ABCB1 expression corrected for differences in vector expression by normalizing ABCB1 median fluorescence intensity (MFI) to EYFP MFI. (B) ABCB1 MFI without correction. The bars represent the means of three replicate measurements ± standard error of mean. Differences in ABCB1 expression were analyzed using Student’s independent t-tests comparing all cell lines to K562/ABCB1 TTT.
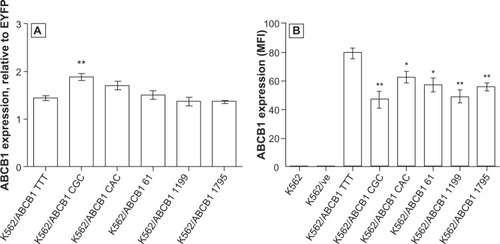
Figure 3 The absence of ABCB1 expression in parental K562 cells. Parental K562 and K562 control cells transduced with an empty vector (K562/ve) were fluorescently labeled with anti-ABCB1 (K562+ and K562/ve +) and analyzed by flow cytometry. Labeled cells had similar fluorescent profiles as unlabeled parental K562 (K562 −), indicating that parental K562 and control cells do not express ABCB1. The figure is a representative result from one of three replicate measurements.
Exposing cells to the ABCB1 substrate vincristine and assessing cell survival using the MTT assay evaluated functionality of transduced ABCB1. It was shown that K562/ABCB1 TTT treated with vincristine had a 17-fold increased IC50 value compared to K562/ve ( and ), indicating a functional ABCB1-transport protein.
Figure 4 The effect of ABCB1 variant haplotypes on drug efficacies. Parental K562, K562 transduced with empty vector (K562/ve), and K562/ABCB1 variant cell lines were exposed to serial dilutions of drugs for 72 hours, followed by analysis of cell survival. Half-maximal inhibitory concentration (IC50) values were derived from each of nine replicate dilution series (vincristine, n = 6), and mean IC50 values are plotted (•), together with error bars representing ±95% confidence intervals (CIs).
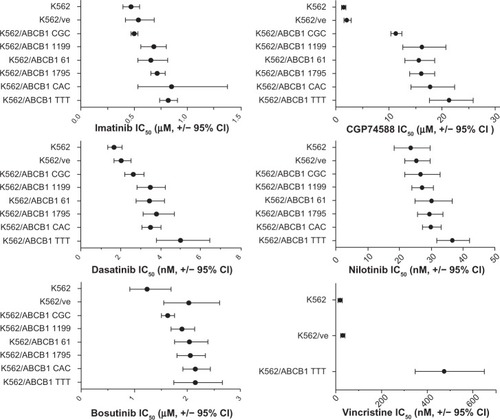
Table 2 Influence of ABCB1 variant haplotypes on drug efficacy
ABCB1 expression mainly influences the efficacy of CGP74588 and dasatinib
The cells that expressed the ABCB1 TTT variant haplotype had the highest IC50 value of all cell lines when treated with any of the TKIs, with the exception of imatinib, where the K562/ABCB1 CAC had a slightly higher IC50 but with a wide confidence interval. The fold change in IC50 of ABCB1 TTT, compared to the IC50 of K562/ve control cells, was used for the comparative evaluation of ABCB1 protection against cytotoxicity of the investigated TKIs. ABCB1 expression seemed to have the largest impact on CGP74588: the IC50 value was ten times higher in K562/ABCB1 TTT than K562/ve. A 2.4-fold increase in IC50 was seen after treating K562/ABCB1 TTT with dasatinib. Compared to K562/ve, K562/ABCB1 TTT had a 1.5-fold increase in IC50 when treated with imatinib and a 1.4-fold increase when treated with nilotinib. Bosutinib efficacy was not affected by ABCB1 expression when comparing the IC50 obtained in ABCB1-expressing cells to that of K562/ve ( and ). However, the K562/ve cells appeared to be more resistant to bosutinib than parental K562, although the 95% confidence intervals of these IC50 values overlapped ().
ABCB1 variants do not influence the efficacy of TKIs
No prominent effects of ABCB1 variants on TKI resistance were detected in this study. The IC50 values obtained when treating cells with the two drugs (dasatinib and CGP74588) most affected by ABCB1 expression showed that only K562/ABCB1 CGC had reduced resistance to the drugs compared to cells that expressed ABCB1 TTT (). However, the K562/ABCB1 CGC cells also had the lowest EYFP and ABCB1 expression compared to other cell lines ( and ). All other ABCB1 variant cell lines had similar resistance to dasatinib and CGP74588 as K562/ABCB1 TTT. The protective effect of ABCB1 expression was small using imatinib and nilotinib and nonsignificant for bosutinib. Similar to CGP74588 and dasatinib, a reduced protective effect of the ABCB1 CGC variant was observed when treating the cells with imatinib. No influence of other genetic variants was detected on the efficacy of these drugs.
ABCB1 expression leads to low intracellular accumulation of dasatinib and CGP74588
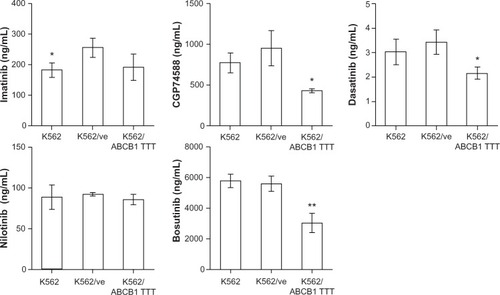
As expected from the results of the MTT assays, K562/ABCB1 TTT accumulated significantly lower quantities of CGP74588 and dasatinib than did K562/ve (). No significant influence of ABCB1 expression on accumulation of imatinib or nilotinib was detected, although K562 accumulated a smaller quantity of imatinib than K562/ve. Surprisingly, there was also a significantly lower accumulation of bosutinib in K562/ABCB1 TTT than K562/ve.
Figure 5 The influence of ABCB1 expression on intracellular accumulation of tyrosine kinase inhibitors (TKIs). Parental K562, K562 transduced with empty vector (K562/ve), and ABCB1 TTT haplotype (K562/ABCB1 TTT) were incubated with TKIs, and intracellular drug accumulation was quantified. The bars represent the mean concentrations of triplicate incubations, with error bars corresponding to ± standard deviation. Differences in drug accumulation were analyzed using Student’s independent t-tests, comparing K562 and K562/ABCB1 TTT to K562/ve.
Discussion
The therapeutic efficacy of drugs, including TKIs used in the treatment of CML, that rely on the long-term effect of steady-state plasma concentrations might be particularly sensitive to alterations in such pharmacokinetic parameters as drug-transport function. But despite substantial efforts, it has proven difficult to establish an association of common ABCB1 SNPs with imatinib pharmacokinetics and outcome in vivo. We report that none of the ABCB1 haplotypes investigated in this study had any major influence on the efficacy of TKIs in K562 cells.
ABCB1 expression provided substantial protection against the cytotoxic effects of CGP74588 and dasatinib. These findings were also supported by reduced intracellular accumulation of CGP74588 and dasatinib in ABCB1-expressing cells compared to the K562/ve control cells. The influence of ABCB1 on imatinib and nilotinib efficacy was minor, albeit significant. These results were in agreement with previous findings showing that ABCB1 expression provides better protection against dasatinib than imatinib or nilotinib cytotoxicity.Citation3,Citation4 The observed resistance of ABCB1-expressing cells to CGP74588 is also in agreement with previous findings showing that CGP74588 is extensively transported in a multidrug-resistant cell line that expresses ABCB1.Citation6 K562/ve cells were equally as sensitive to bosutinib cytotoxicity as ABCB1-expressing cells, but appeared to be more resistant than the parental K562 cells. However, ABCB1 expression significantly reduced the intracellular quantity of bosutinib. Based on these data, an influence of ABCB1 expression on bosutinib transport cannot be ruled out, even though this was not reported by previous investigators.Citation4
Plasma concentrations of TKIs measured in patients at standard dosing reach approximately 1.7 μM of imatinib,Citation13 0.4 μM of CGP74588,Citation13 2.1 μM of dasatinib,Citation32 4.2 μM of nilotinib,Citation33 and 0.4 μM of bosutinib.Citation34 The drug concentrations studied here were well within clinically attainable plasma concentrations of imatinib, dasatinib, and nilotinib. CGP74588 and bosutinib concentrations were higher than the mean observations in vivo. However, considering the range of individual variation in plasma concentrations of CGP74588 and bosutinib in vivo,Citation13,Citation34 the concentrations used in the present study can be considered as clinically attainable.
A comparison of the efficacies of TKIs and the previously validated ABCB1 substrate vincristine in K562/ABCB1 TTT revealed that imatinib, dasatinib, and nilotinib should all be regarded as rather poor ABCB1 substrates. The K562/ABCB1 TTT had only 1.4- to 2.4-fold better protection against the toxicity of these TKIs, compared to the 17-fold protection observed when using vincristine. Only CGP74588 may be regarded as a relatively good substrate of ABCB1, given its tenfold-higher IC50 in K562/ABCB1 TTT compared to K562/ve. These data suggest that CGP74588-should be affected by ABCB1-transport activity to a much larger extent than imatinib in vivo. This could be important to keep in mind when interpreting association studies of ABCB1 function and therapeutic efficacy of imatinib, considering that although this metabolite is less potent than imatinib, it might accumulate in patients with low ABCB1-transport activity. We previously found an inverse association of CYP3A metabolic activity and imatinib therapeutic outcome, indicating the possible significance of imatinib metabolites in CML therapy.Citation8
Evaluations of ABCB1 haplotypes and their influence on protection against TKI cytotoxicity revealed that only the CGC haplotype significantly reduced cellular resistance to these drugs compared to the ABCB1 TTT haplotype. However, K562/ABCB1 CGC had the lowest EYFP expression among the transduced cell lines, indicating less transcriptional activity of the vector that slightly reduced ABCB1 expression compared to other cell lines. Therefore, the reduced resistance of K562/ABCB1 CGC might be an effect of lower transcriptional activity in this particular cell line rather than of the variant haplotype. None of the other investigated ABCB1 variant haplotypes had any significant influence on either ABCB1 membrane expression or TKI efficacy. It was concluded that neither of the investigated ABCB1 haplotypes were likely to influence TKI transport. Our findings were in agreement with other in vitro studies that did not find any association of ABCB1 genotypes and transport functions.Citation35,Citation36 Considering the chosen method of retroviral gene transfer, it was not possible to investigate the influence of ABCB1 SNPs on pretranslational mechanisms such as messenger RNA stability. It has previously been suggested that the 3435T>C SNP affects ABCB1 messenger RNA levels,Citation37 which our data could neither confirm nor refute.
We have shown that investigated ABCB1 haplotypes do not influence the efficacy of TKIs in vitro, at least on a posttranslation level; this is supported by the negative explorations in several association studies of the ABCB1 genotype and outcome of imatinib in vivo.Citation22–Citation25 Given our results that TKIs currently used in CML therapy (imatinib, dasatinib, and nilotinib) appear to be rather poor ABCB1 substrates compared to traditional chemotherapeutic agents, a large impact on TKI efficacy from varying ABCB1 activity is perhaps not to be expected. Our results indicate that the most common ABCB1 SNPs are not likely to predict response and resistance to TKI therapy in vivo. However, there is still a possibility that other ABCB1 SNPs or haplotypes not yet investigated have a greater impact on ABCB1-transport function than those found here. According to our findings, dasatinib and the major metabolite of imatinib – CGP74588 – would be among the compounds most likely to be affected.
All TKIs studied here are subjected to additional sources of pharmacokinetic variability. It is known that ABCB1 and CYP3A4 to some degree have overlapping substrate specificities and tissue distributions.Citation38 Indeed, all TKIs used in the present study were CYP3A4 substratesCitation39,Citation40 and might be influenced by variations in CYP3A4 metabolic activity, but also by other sources of variation, such as degree of plasma protein binding and the variable uptake and efflux by other transport proteins. To some extent, the outcome of imatinib can be predicted by the activity of the OCT-1 uptake transporter,Citation15 while dasatinib and nilotinib do not seem to be OCT-1 substrates.Citation5,Citation41 In addition, all TKIs except bosutinib are transported by ABCG2,Citation3,Citation4 which is coexpressed with ABCB1 in the liverCitation42 and in the CML stem cells,Citation11,Citation43 potentially influencing the distribution and elimination of TKIs. Further studies on imatinib as well as the second-generation TKIs are needed in order to understand fully the rate-limiting steps of TKI pharmacokinetics, any potential additive effects of the different pharmacokinetic parameters, and their significance as predictors of response and resistance to the TKIs used in CML therapy.
Conclusion
ABCB1 expression substantially influenced the transport and efficacy of dasatinib and the imatinib metabolite CGP74588, which is a far better substrate for ABCB1 than the parent compound. None of the investigated ABCB1 variant haplotypes influenced the efficacy of TKIs used in CML therapy. This result indicates that any influence of ABCB1-transport activity on TKI efficacy in vivo is not limited to non-synonymous SNPs, but might involve other regulatory elements of ABCB1 activity in addition to SNPs or haplotypes not yet investigated.
Acknowledgments
The authors would like to thank the Swedish Research Council, the Swedish Cancer Society, the Medical Research Council of Southeast Sweden and the Linköping University Cancer Research Network for funding this study. We are also grateful to Novartis Pharma AG for providing imatinib and CGP74588.
Supplementary materials
Method for quantification of intracellular TKI accumulation
K562, K562/ve, and K562/ABCB1 TTT were incubated with TKIs, followed by quantification of intracellular drug concentrations using a method modified from a previous report.Citation1 Two methods were developed: one for quantification of imatinib, CGP74588, and bosutinib where dasatinib (800 ng/mL) served as the internal standard, and another for dasatinib and nilotinib, using imatinib (80 ng/mL) as the internal standard.
Cells were seeded 400,000/mL in 5 mL of growth medium and incubated for 120 minutes (imatinib, CGP74588, or bosutinib) or 180 minutes (dasatinib or nilotinib), depending on the time to attain influx–efflux equilibrium. TKIs were used in concentrations in the same range as mean IC50 concentrations found in parental K562 cells using the MTT assay (imatinib 0.5 μM, CGP74588 2.0 μM, dasatinib 1.5 nM, nilotinib 20 nM, bosutinib 2.0 μM). Cells were separated from the medium by centrifugation (4,000 g, 5 minutes at 22°C) on 1.5 mL silicone oil. Cell pellets were disrupted by adding 200 μL of internal standard in 4% formic acid (FA) in water (v/v), with the exception of pellets incubated with dasatinib or nilotinib, which were disrupted using 100 μL of 4% FA. Lysates were centrifuged at 10,000 g for 10 minutes at 4°C, and supernatants were collected and diluted 1:10 in water before analysis, with the exception of extracts from dasatinib and nilotinib incubations, which were analyzed as concentrates.
Samples were analyzed on a chromatographic system (Acquity UPLC System; Waters, Milford, MA, USA) coupled to the tandem-quadrupole mass spectrometer Xevo TQ MS (Waters). Five microliters of samples were separated on an Acquity UPLC BEH C18 (2.1 × 50 mm, 1.7 μm) column (Waters), using a gradient mobile phase of 0.1% FA (v/v) in water (A) and 0.1% FA (v/v) in acetonitrile (B). A gradient was delivered at 0.6 mL/minute − 0.0–0.4 minutes, 80% A; 0.4–3.0 minutes, linear gradient to 20% A; 3.0–3.5 minutes, 20% A – followed by reequilibration with 80% A to 4.0 minutes. Multiple-reaction monitoring was applied and TKIs were monitored at transitions m/z 494 > 394 for imatinib, 480 > 394 for CGP74588, 488 > 232 and 488 > 401 for dasatinib, 530 > 289 for nilotinib, and 530 > 141 for bosutinib.
Calibrators were prepared in blank lysates and extracted in accordance with the same procedure as for the samples. The calibration curve ranges were 10–3,000 ng/mL for imatinib, CGP7488 and bosutinib; 1–500 ng/mL for dasatinib; and 25–500 ng/mL for nilotinib. Quality-control samples were prepared in two concentrations in blank lysates for each calibration curve: imatinib, CGP74588, and bosutinib were analyzed at 100 ng/mL and 2,500 ng/mL; dasatinib at 8 ng/mL and 200 ng/mL; and nilotinib at 30 ng/mL and 200 ng/mL.
The calibration curves were used for calculation of the TKI concentration in samples and normalized to the internal standard. All compounds had assay imprecision < 10%, with accuracy ranging from 85% to 113% at the investigated quality-control concentrations (n = 5).
Reference
- MlejnekPNovakODolezelPA non-radioactive assay for precise determination of intracellular levels of imatinib and its main metabolite in Bcr-Abl positive cellsTalanta2011831466147121238737
Disclosure
The authors report no conflicts of interest in this work.
References
- KantarjianHShahNPHochhausADasatinib versus imatinib in newly diagnosed chronic-phase chronic myeloid leukemiaN Engl J Med20103622260227020525995
- KantarjianHMHochhausASaglioGNilotinib versus imatinib for the treatment of patients with newly diagnosed chronic phase, Philadelphia chromosome-positive, chronic myeloid leukaemia: 24-month minimum follow-up of the phase 3 randomised ENESTnd trialLancet Oncol20111284185121856226
- DohseMScharenbergCShuklaSComparison of ATP-binding cassette transporter interactions with the tyrosine kinase inhibitors imatinib, nilotinib, and dasatinibDrug Metab Dispos2010381371138020423956
- HegedusCOzvegy-LaczkaCApátiAInteraction of nilotinib, dasatinib and bosutinib with ABCB1 and ABCG2: implications for altered anti-cancer effects and pharmacological propertiesBr J Pharmacol20091581153116419785662
- HiwaseDKSaundersVHewettDDasatinib cellular uptake and efflux in chronic myeloid leukemia cells: therapeutic implicationsClin Cancer Res2008143881388818559609
- MlejnekPDolezelPFaberEKosztyuPInteractions of N-desmethyl imatinib, an active metabolite of imatinib, with P-glycoprotein in human leukemia cellsAnn Hematol20119083784221225261
- CohenMHWilliamsGJohnsonJRApproval summary for imatinib mesylate capsules in the treatment of chronic myelogenous leukemiaClin Cancer Res2002893594212006504
- GreenHSkoglundKRommelFMirghaniRALotfiKCYP3A activity influences imatinib response in patients with chronic myeloid leukemia: a pilot study on in vivo CYP3A activityEur J Clin Pharmacol20106638338620054526
- ThiebautFTsuruoTHamadaHGottesmanMMPastanIWillinghamMCCellular localization of the multidrug-resistance gene product P-glycoprotein in normal human tissuesProc Natl Acad Sci U S A198784773577382444983
- ThomasJWangLClarkREPirmohamedMActive transport of imatinib into and out of cells: implications for drug resistanceBlood20041043739374515315971
- JiangXZhaoYSmithCChronic myeloid leukemia stem cells possess multiple unique features of resistance to BCR-ABL targeted therapiesLeukemia20072192693517330101
- RacilZRazgaFPolakovaKMAssessment of adenosine triphosphate-binding cassette subfamily B member 1 (ABCB1) mRNA expression in patients with de novo chronic myelogenous leukemia: the role of different cell typesLeuk Lymphoma20115233133421133723
- LarsonRADrukerBJGuilhotFImatinib pharmacokinetics and its correlation with response and safety in chronic-phase chronic myeloid leukemia: a subanalysis of the IRIS studyBlood20081114022402818256322
- PicardSTitierKEtienneGTrough imatinib plasma levels are associated with both cytogenetic and molecular responses to standard-dose imatinib in chronic myeloid leukemiaBlood20071093496349917192396
- WhiteDLRadichJSoveriniSChronic phase chronic myeloid leukemia patients with low OCT-1 activity randomized to high-dose imatinib achieve better responses and have lower failure rates than those randomized to standard-dose imatinibHaematologica20129790791422207690
- CascorbiIP-glycoprotein: tissue distribution, substrates, and functional consequences of genetic variationsHandb Exp Pharmacol201120126128321103972
- KroetzDLPauli-MagnusCHodgesLMSequence diversity and haplotype structure in the human ABCB1 (MDR1, multidrug resistance transporter) genePharmacogenetics20031348149412893986
- DeenikWvan der HoltBJanssenJJPolymorphisms in the multidrug resistance gene MDR1 (ABCB1) predict for molecular resistance in patients with newly diagnosed chronic myeloid leukemia receiving highdose imatinibBlood201011661446145 author reply 6145–614621183698
- DulucqSBouchetSTurcqBMultidrug resistance gene (MDR1) polymorphisms are associated with major molecular responses to standard-dose imatinib in chronic myeloid leukemiaBlood20081122024202718524988
- NiLNLiJYMiaoKRMultidrug resistance gene (MDR1) polymorphisms correlate with imatinib response in chronic myeloid leukemiaMed Oncol20112826526920204543
- GurneyHWongMBalleineRLImatinib disposition and ABCB1 (MDR1, P-glycoprotein) genotypeClin Pharmacol Ther200782334017495881
- MarinDBazeosAMahonFXAdherence is the critical factor for achieving molecular responses in patients with chronic myeloid leukemia who achieve complete cytogenetic responses on imatinibJ Clin Oncol2010282381238820385986
- TakahashiNMiuraMScottSAInfluence of CYP3A5 and drug transporter polymorphisms on imatinib trough concentration and clinical response among patients with chronic phase chronic myeloid leukemiaJ Hum Genet20105573173720720558
- KimDHSriharshaLXuWClinical relevance of a pharmacogenetic approach using multiple candidate genes to predict response and resistance to imatinib therapy in chronic myeloid leukemiaClin Cancer Res2009154750475819584153
- SeongSJLimMSohnSKInfluence of enzyme and transporter polymorphisms on trough imatinib concentration and clinical response in chronic myeloid leukemia patientsAnn Oncol20122475676023117072
- GréenHFalkIJLotfiKAssociation of ABCB1 polymorphisms with survival and in vitro cytotoxicty in de novo acute myeloid leukemia with normal karyotypePharmacogenomics J20121211111820938465
- GreenHSoderkvistPRosenbergPHorvathGPetersonCABCB1 G1199A polymorphism and ovarian cancer response to paclitaxelJ Pharm Sci2008972045204817828752
- CascorbiIGerloffTJohneAFrequency of single nucleotide polymorphisms in the P-glycoprotein drug transporter MDR1 gene in white subjectsClin Pharmacol Ther20016916917411240981
- DeKoterRPSchweitzerBLKamathMBRegulation of the interleukin-7 receptor alpha promoter by the Ets transcription factors PU.1 and GA-binding protein in developing B cellsJ Biol Chem2007282141941420417392277
- MosmannTRapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assaysJ Immunol Methods19836555636606682
- MarzoliniCPausEBuclinTKimRBPolymorphisms in human MDR1 (P-glycoprotein): recent advances and clinical relevanceClin Pharmacol Ther200475133314749689
- ChristopherLJCuiDWuCMetabolism and disposition of dasatinib after oral administration to humansDrug Metab Dispos2008361357136418420784
- NovartisNDA 22-068: Clinical pharmacology and biopharmaceutics review2006 Available from: http://www.accessdata.fda.gov/drugsatfda_docs/nda/2007/022068s000_ClinPharmR.pdfAccessed April 29, 2013
- HsyuPHMouldDRUptonRNAmanteaMPharmacokinetic-pharmacodynamic relationship of bosutinib in patients with chronic phase chronic myeloid leukemiaCancer Chemother Pharmacol20137120921823070145
- Kimchi-SarfatyCGribarJJGottesmanMMFunctional characterization of coding polymorphisms in the human MDR1 gene using a vaccinia virus expression systemMol Pharmacol2002621612065748
- MoritaNYasumoriTNakayamaKHuman MDR1 polymorphism: G2677T/A and C3435T have no effect on MDR1 transport activitiesBiochem Pharmacol2003651843185212781336
- HoffmeyerSBurkOvon RichterOFunctional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivoProc Natl Acad Sci U S A2000973473347810716719
- WacherVJWuCYBenetLZOverlapping substrate specificities and tissue distribution of cytochrome P450 3A and P-glycoprotein: implications for drug delivery and activity in cancer chemotherapyMol Carcinog1995131291347619215
- van ErpNPGelderblomHGuchelaarHJClinical pharmacokinetics of tyrosine kinase inhibitorsCancer Treat Rev20093569270619733976
- AbbasRHugBALeisterCBurnsJSonnichsenDEffect of ketoconazole on the pharmacokinetics of oral bosutinib in healthy subjectsJ Clin Pharmacol2011511721172721148045
- WhiteDLSaundersVADangPOCT-1-mediated influx is a key determinant of the intracellular uptake of imatinib but not nilotinib (AMN107): reduced OCT-1 activity is the cause of low in vitro sensitivity to imatinibBlood200610869770416597591
- MaliepaardMSchefferGLFaneyteIFSubcellular localization and distribution of the breast cancer resistance protein transporter in normal human tissuesCancer Res2001613458346411309308
- JordanidesNEJorgensenHGHolyoakeTLMountfordJCFunctional ABCG2 is overexpressed on primary CML CD34+ cells and is inhibited by imatinib mesylateBlood20061081370137316627755
- National Center for Biotechnology Information 1000 Genomes Browser version 3.0 Available from: http://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/Accessed August 1, 2013