Abstract
Crohn’s disease is an immune-related disorder characterized by inflammation of the gastrointestinal mucosa, which can occur in any area throughout the digestive tract. This life-long disease commonly presents with abdominal pain, diarrhea, vomiting, and weight loss. While the exact etiology of this disease is largely unknown, it is thought to arise from an interaction between microbial, immunological, and environmental factors in a genetically susceptible host, whereby the immune system attacks the intestine as it cross reacts against gut microbial antigens. The study of genetic variants associated with Crohn’s disease has shed light on our understanding of disease pathophysiology. A large number of genetic variants identified in Crohn’s disease are related to genes targeting microbial recognition and bacterial wall sensing, the most common being NOD2/CARD15 gene. This review will discuss the recent advance in our knowledge of genetic variants of this disease and how they influence the disease course and prognosis.
Keywords:
Introduction
Inflammatory bowel disease (IBD) is defined as a group of chronic intestinal conditions characterized by inflammation in the intestine. The most common types of IBD are Crohn’s disease (CD) and ulcerative colitis (UC). CD can occur in any area along the alimentary tract, but UC is localized primarily to the large intestine. Full thickness of the bowel wall is often affected in CD, while only the mucosa is involved in UC. IBD is a common disorder worldwide, with a prevalence of 0.1%–0.4% of the general population.Citation1 In the United States, the Center for Disease Control estimates that over 1.4 million Americans are affected by this disease, with an overall estimated annual health care cost exceeding US$1.7 billion (cdc.gov/ibd/). IBD is currently without a cure, requires life-long care, interferes with quality of life, and frequently necessitates surgical intervention. Therefore, it has been recognized as one of the top five gastrointestinal disease burdens in the USA. Recent international trends suggest that there is an increase in the incidence of CD.Citation2 Whether this finding represents a true rise in the incidence of the disease or merely reflects an improvement in diagnostic and reporting methods is difficult to determine at the present time. The past few decades have heralded new discoveries in IBD, most of which are related to the genetics of the disease, diagnostic tools, and management strategies. Recent discoveries have brought much attention to the genetic predisposition of patients with IBD. Those genetic variations are largely related to the innate immune system and microbial recognition.Citation3–Citation7
Genetic variants in Crohn’s disease
CD is a complex, multifactorial disease with strong genetic associations that lead to aberrant, chronic mucosal inflammation after exposure to environmental or microbial triggers. Thus far, 163 risk loci have been identified in patients with IBD,Citation8 many of which are involved in innate immune responses and mucosal homeostasis. Currently, our genetic knowledge of CD is applied to understanding the natural history of the disease, describing racial predominance, predicting early onset stricturing and fistulizing, and determining location and overall severity of disease. Wagner et al recently described an association between mutations in toll-like receptor (TLR) 4 (rs4986790) and interleukin 10 receptor, alpha subunit (rs22291130) with Mycobacterium avium subspecies paratuberculosis positive CD patients,Citation9 further implicating the role of microbial triggers and aberrant genetic responses in the immune system. This genetic data is helpful information that will possibly lead to the ultimate discovery of etiologies for the disease.
Significant CD risk has been associated with NOD2/CARD15,Citation10 IBD5, and DLG5;Citation11 IL23R and ATG16L1Citation4 have also been identified as CD susceptibility genes (see for gene expansions). Genetic variants of Immunity-related GTPase family M (IRGM) have been significantly associated with CD,Citation5 and it will be discussed later in this review, along with the other variant in the autophagy pathway, ATG16L1.
Table 1 Most clinically relevant and GWAS replicated CD genes
Three single nucleotide polymorphisms (SNPs) found within the NOD2/CARD15 gene (R702W, G908R, and 1007fsinsC) were found to be racially distinct. These SNPs have been established as independent risk factors for CD in Caucasians.Citation10,Citation12–Citation14 Biank et al reviewed ten manuscripts, which together included 1319 children with NOD2/CARD15 mutations.Citation15 They concluded that Caucasian children with European ancestry have similar attributable risks for developing CD as adults due to NOD2/CARD15. Furthermore, as stated previously, and currently restated by Biank et al,Citation15 the NOD2/CARD15 mutational variants are nearly absent or rare in East Asians, Arabs, Africans, and African-Americans.Citation16–Citation20
Genome wide association studies (GWAS) have been used very effectively in IBD genetic research. Deep resequencing of GWAS data has identified highly significant variants at CARD9, NOD2/CARD15, CUL2, and IL18RAP, which contribute to risk associations of previously defined variants at these loci, and as well, showed the functionality of the newly implicated NOD2/CARD15 variants. Rivas et al went on to describe additional protective variants at IL23R,Citation21 and Stoll et alCitation22 showed DLG5 R30Q is a modest risk gene for CD.
Further analysis of the data by Stoll et al revealed gender differences in allele frequency of R30Q between adult male CD cases and controls.Citation22 They concluded that the DLG5 R30Q variant confers increased risk to adult males for CD. Biank et alCitation15 compared the results of their pediatric CD study sample to the adult CD study sample results reported by Friedrichs et alCitation23 and found the allele frequency of R30Q in children with CD to be 10.7% for males and 5.6% for females. The allele frequency of R30Q for controls was 8.3% for males and 12.3% for females. This difference of allele frequencies among females with CD and controls shows that R30Q may have a protective effect in female children.Citation15
Innate cytokines produced by antigen presenting cells in response to microbial stimulation are critical for the induction of specific T effector responses that can combat invading bacteria or viruses. Interleukin (IL)-12 is an innate cytokine composed of a p40 and a p35 subunits, which are essential for the polarization of interferon-γ-producing T helper 1 (TH1) cells. Interestingly a second innate cytokine called IL-23 was discovered in 2000,Citation24 and it shares the p40 subunit of IL-12, but instead of p35, it heterodimerizes with a unique p19 chain. Early studies examining IL-12 deletion of the p40 chain or via neutralization by antibodies targeting the p40 chain may have also effected IL-23-mediated responses.Citation25,Citation26 However, over the last 12 years IL-23 has emerged as a proinflammatory driver in gastrointestinal inflammation. IL-12 is important for TH1 and interferon-γ responses, whereas IL-23 has been shown to expand IL-17 producing TH17 cellsCitation27 and IL-17-producing innate lymphoid cellsCitation28 via expression of IL-23R.Citation28,Citation29
The IL-23/IL-17 inflammatory axis has now been implicated in the pathogenesis of Crohn’s disease.Citation30–Citation32 This conclusion comes in part from the findings that CD patients have a higher number of circulating IL-17+ cellsCitation33 and from a study by Rismo et al,Citation34 which showed that increased messenger (m) RNA expression of tumor necrosis factor (TNF) and IL17A in healed mucosa significantly increased the risk of relapse (hazard ratio [HR] = 3.4, P = 0.03, sensitivity 80%, specificity 38% and HR = 4.1, P = 0.008, sensitivity 81%, specificity 61%, respectively).
More thorough investigations linking genetic variants of IL23, IL17, NOD2/CARD15, IRGM, ATG16L1, and DLG5 to mRNA expression, protein expression, gender, age, race, and phenotypic outcomes will help us to further understand the etiologies of Crohn’s disease. A comprehensive list of the main genetic variants in CD is summarized in . In summary, the mutational variants in CD further describe a defect in bacterial sensing, as mentioned by Wagner et al,Citation9 and an exaggerated response of the intestinal immune system.
Autophagy in Crohn’s disease
Autophagy is a conserved cellular pathway that maintains cellular homeostasis via the degradation of cytosolic contents and proteins.Citation35 This process occurs via the sequestration of cytoplasmic contents into autophagosomes and the subsequent fusion with lysosomes for degradation and recycling ().Citation36,Citation37 Autophagy has drawn interest beyond the extent of homeostatic regulation and is now a widely recognized pathophysiological process in autoimmune diseases,Citation38–Citation46 cancer,Citation47,Citation48 and IBD.Citation3,Citation49,Citation50
Figure 1 Genes of the autophagy pathway implicated in Crohn’s disease susceptibility.
Abbreviations: LRRK2, leucine-rich repeat kinase 2; IRGM, immunity-related GTPase family M protein; NOD2, nucleotide-binding oligomerization domain-containing protein 2; CARD15, caspase recruitment domain-containing protein 15; ATG16L1, autophagy-related protein 16–1; GTP, guanosine triphosphate; ULK1, serine/threonine kinase.
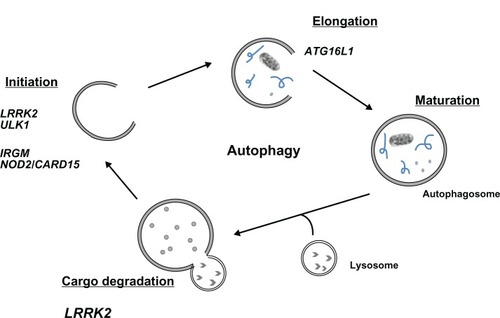
Intestinal tissues are in close proximity to the microbial world and therefore rely on intracellular defense mechanisms like autophagy in order to prevent aberrant or harmful inflammation. Thus, defects in directing commensals and pathogens to the autophagy pathway may impair bacterial responses and promote an environment in which anticommensal immunity and inflammation is favored.
ATG16L1 gene variant rs2241880 (T300A) is a non-synonomous coding SNP that results in a single amino acid change in the mature protein. rs2241880 was identified in both a transmission disequilibrium test and a case controlled comparison in a study by Hampe et alCitation4 examining CD patients in a German cohort. These data suggest that the CD risk conferred by ATG16L1 gene variation is confined to individuals carrying allele G, with a 60% frequency of the G allele in affected individuals compared to 53% in controls. These data were confirmed in patients from the United Kingdom (G allele frequency was 59% affected versus 52% in controls).Citation4 Interestingly, a statistical interaction between rs2241880 and the CARD15 genotypes was discovered.Citation4 This genetic association is further supported by the findings that muramyl dipeptide enhances autophagy and is inhibited by ATG16L1CD risk allele.Citation51,Citation52 Additionally, the ATG16L1 (T300A) variant has been associated with elevated IL-1β mRNA levels independently of inflammasome activation but not TLR ligands.Citation53 These data suggest that defects in autophagy lead to increased IL-1β secretion and inflammation downstream of NOD2/CARD15.
In addition to activation via pathogen recognition receptors,Citation52,Citation54,Citation55 type 1 and type II interferons are potent inducers of the autophagy pathway.Citation56–Citation58 Interferon-γ can induce the expression of immunity-related guanosine triphosphate (IRG) proteins in mice while human IRG lack interferon response elements. IRGM is the most well-known member of the IRG family and is constitutively expressed at high levels in human cells.Citation56 Similar to ATG16L1, genetic variants of IRGM have been significantly associated with CD risk;Citation5,Citation59 however, unlike ATG16L1, studies have been unable to correlate NOD2/CARD15 variants with IRGM.Citation60,Citation61
Interestingly the lead SNP in IRGM (rs13361189) identified in CD is in perfect linkage disequilibrium with a 20 kb deletion upstream to the IRGM locus,Citation59 hence the risk haplotype includes both deletion and SNP. Brest et alCitation62 demonstrated that rs13361189 and the deletion are in perfect linkage disequilibrium with a synonymous exonic SNP (rs10065172) that alters IRGM levels and the binding of microRNA196.Citation62 IRGM knockdown and microRNA196 over-expression prevents the targeting of adherent invasive E. coli to the autophagosome,Citation62 suggesting that aberrant immune activity or loss of tolerance to commensal may explain the role of IRGM rs13361189 in CD.
While ATG16L1 is the most widely examined and well-characterized variant of the autophagy pathway associated with CD, recent studies have identified other important genes. Leucine-rich repeat kinase 2 (LRRK2) is a member of the leucine-rich repeat kinase family.Citation63 Three studies have identified variants in LRRK2 associated with CD.Citation64–Citation66 The SNP, rs11175593, is located in a noncoding region on chromosome 12. Interestingly, reports have shown that LRRK2 may be a factor in autophagy via a scaffolding role during protein-protein interaction.Citation67–Citation69 ULK1 is a serine/threonine-protein kinase, which forms a complex with Atg1 and activates the Atg1/ULK1 complex via dephosphorylation of Atg1. This is the most upstream step of autophagosome formation.Citation70 The SNP in ULK1 (rs12303764) was identified by Henckaerts et alCitation71 in a population study of 1282 CD patients of Western European origin. NOD2/CARD15 variants were shown to have no impact on the association of rs12303764 with CD.Citation71 The identification of SNPs in LRRK2 and ULK1, and the already described role of ATG16L1, further establish the regulation of autophagy as an important cellular function in CD. However, more in-depth analysis of both LRRK2 and ULK1 SNPs must be performed to fully understand the functional consequence in CD pathogenesis.
Genetic predictors of Crohn’s disease clinical outcome
Since genetic factors do not change over time, the identification of genetic variants that may predict disease behavior and prognosis are superior to other predictive factors such as serological and clinical parameters. Despite the ever-growing number of susceptibility loci in CD, there are only a few variants associated with statistically significant clinical outcome and prognosis.Citation72 In addition to its identification as a susceptibility locus, NOD2/D15 has also been identified in studies examining outcomes of CD. The presence of NOD2/CARD15 has been associated with a more aggressive clinical course involving higher risk of intestinal strictures, earlier need for surgical intervention, and less postoperative disease-free intervals.Citation13,Citation73,Citation74 NOD2/CARD15 was also found to be the most important factor for ileal location and stenosing and penetrating disease.Citation72
The presence of a genetic variant in Janus kinase 2 (JAK2) as a predictor for ileal involvement and stenosing disease was found in a cohort of 1528 European CD patients.Citation72 Interestingly Prager et alCitation75 demonstrated that patients with the C risk allele within JAK2 rs10758669 have increased epithelial permeability, suggesting that the effect on disease behavior may be due to alterations in overall intestinal permeability.
Other genetic variants that impact disease behavior include an SNP in IRGM (rs4958847), which was significantly associated with frequency of surgery in a study of 66 patients with ileocecal CD, and a negative association was found between ileal disease and TLR1 S602I.Citation76 Genetic variants may also impact the response to certain immune therapies; polymorphisms in multidrug resistant 1, TNF, and migration inhibitory factor genes have all been associated with sensitivity to corticosteroid,Citation77,Citation78 while variants in apoptosis genes have been used to predict the response to infliximab therapy.Citation79 The identification of genetic variants could be an important tool in predicting disease behavior and responsiveness of certain patients to specific treatments; however, this may be complicated by the major role that other factors, such as the environment, plays in CD pathogenesis. It may be worthwhile to examine genetic variants alongside clinical, serological, and microbiological data in order to more accurately predict disease behavior.
Genetic variants in pediatric Crohn’s disease
Phenotypic differences between adult and pediatric onset CD suggest a different genetic predisposition. Pediatric IBD is characterized by extensive intestinal involvement and rapid early progression.Citation80 The increased risk of CD among family members with early-onset disease and the stratification of disease by age of onset has been the focus of many studies in the effort to further ascertain the genetic influence of CD.
Genetic susceptibility may play a more important role in the etiology of early-onset IBD than in late-onset IBD, and therefore pediatric-onset IBD patients can be expected to have a higher frequency of gene mutations. De Ridder and colleaguesCitation81 investigated genetic polymorphism in CARD15 and DLG5 and described more frequent occurrence of polymorphisms, 3020insC, in CARD15 and SNP, rs3792876, in SLC22A4/5 in patients with pediatric-onset CD than in patients with adult-onset CD. Polymorphisms 3020insC in CARD15 and SNP rs2165047 in DLG5 were associated with specific phenotypes such as ileal and perianal disease, respectively. In another pediatric study in the GWAS literature,Citation82 a previously unreported susceptibility locus for pediatric-onset IBD at 20q13 and 21q22 was identified, suggesting that TNFRSF6B is the most plausible candidate within the 20q13 locus, involved in both antigen-presenting cell differentiation and lymphocyte function. Autophagy-associated genes (ATG16L1 and IRGM) were also seen in early-onset CD cases, further confirming autophagy as an important contributor in the pathogenesis of pediatric CD.Citation83
Dubinsky et alCitation84 examined associations between the R381Q variant of the IL-23R gene and CD in a pediatric cohort and reported significant associations using a family-based study. Similarly, Van Limbergen et alCitation85 also reported associations with the same SNP in a Scottish cohort of pediatric CD using the case-control design. These reports provide strong evidence that the IL-23R gene is not only associated with adult-onset CD, but similar associations exist for children among most Caucasian populations.
Conclusions and perspectives
Currently there are more than 100 independent genetic loci contributing to the pathophysiology of CD. While many of these genes are considered risk factors, we are beginning to discover other genes that may help predict disease behavior as well as the response to certain immune therapies. Particularly interesting will be the further investigation of genes associated with pediatric early onset, thus allowing us the capability to identify children at a higher risk of disease and provide a more effective therapeutic approach. The last decade has witnessed a great expansion in our knowledge of the genetic variants in CD and the identification of many genetic factors that influence the innate immune response. These findings have provided an extensive framework for unraveling the genetic basis of disease susceptibility, clinical predictors of disease behavior, and biological processes of disease. As our understanding becomes more robust, greater insight into the genetic influence in CD will continue to be critical for the development of future therapeutic strategies.
Disclosure
The authors report no conflicts of interests in this work.
References
- MathewCGNew links to the pathogenesis of Crohn disease provided by genome-wide association scansNat Rev Genet20089191417968351
- BenchimolEIGuttmannAToTRabeneckLGriffithsAMChanges to surgical and hospitalization rates of pediatric inflammatory bowel disease in Ontario, Canada (1994–2007)Inflamm Bowel Dis201117102153216121910177
- RiouxJDXavierRJTaylorKDGenome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesisNat Genet200739559660417435756
- HampeJFrankeARosenstielPA genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1Nat Genet200739220721117200669
- ParkesMBarrettJCPrescottNJSequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibilityNat Genet200739783083217554261
- DuerrRHTaylorKDBrantSRA genome-wide association study identifies IL23R as an inflammatory bowel disease geneScience200631458041461146317068223
- TremellingMCummingsFFisherSAIL23R variation determines susceptibility but not disease phenotype in inflammatory bowel diseaseGastroenterology200713251657166417484863
- JostinsLRipkeSWeersmaRKHost-microbe interactions have shaped the genetic architecture of inflammatory bowel diseaseNature2012491742211912423128233
- WagnerJSkinnerNACatto-SmithAGTLR4, IL10RA, and NOD2 mutation in paediatric Crohn’s disease patients: an association with Mycobacterium avium subspecies paratuberculosis and TLR4 and IL10RA expressionMed Microbiol Immunol Epub322013
- HugotJPChamaillardMZoualiHAssociation of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s diseaseNature2001411683759960311385576
- WeersmaRKStokkersPCvan BodegravenAADutch Initiative on Crohn and Colitis (ICC)Molecular prediction of disease risk and severity in a large Dutch Crohn’s disease cohortGut200958338839518824555
- OguraYBonenDKInoharaNA frameshift mutation in NOD2 associated with susceptibility to Crohn’s diseaseNature2001411683760360611385577
- AbreuMTTaylorKDLinYCMutations in NOD2 are associated with fibrostenosing disease in patients with Crohn’s diseaseGastroenterology2002123367968812198692
- AhmadTArmuzziABunceMThe molecular classification of the clinical manifestations of Crohn’s diseaseGastroenterology2002122485486611910336
- BiankVBroeckelUKugathasanSPediatric inflammatory bowel disease: clinical and molecular geneticsInflamm Bowel Dis200713111430143817600381
- LeongRWArmuzziAAhmadTNOD2/CARD15 gene polymorphisms and Crohn’s disease in the Chinese populationAliment Pharmacol Ther200317121465147012823148
- SugimuraMKinouchiYTakahashiSCARD15/NOD2 mutational analysis in Japanese patients with Crohn’s diseaseClin Genet200363216016212630966
- CroucherPJMascherettiSHampeJHaplotype structure and association to Crohn’s disease of CARD15 mutations in two ethnically divergent populationsEur J Hum Genet200311161612529700
- InoueNTamuraKKinouchiYLack of common NOD2 variants in Japanese patients with Crohn’s diseaseGastroenterology20021231869112105836
- YamazakiKTakazoeMTanakaTKazumoriTNakamuraYAbsence of mutation in the NOD2/CARD15 gene among 483 Japanese patients with Crohn’s diseaseJ Hum Genet200247946947212202985
- RivasMABeaudoinMGardetADeep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel diseaseNat Genet201143111066107321983784
- StollMCorneliussenBCostelloCMGenetic variation in DLG5 is associated with inflammatory bowel diseaseNat Genet200436547648015107852
- FriedrichsFBrescianiniSAnneseVEvidence of transmission ratio distortion of DLG5 R30Q variant in general and implication of an association with Crohn disease in menHum Genet2006119330531116446977
- OppmannBLesleyRBlomBNovel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12Immunity200013571572511114383
- NeurathMFFussIKelsallBLStüberEStroberWAntibodies to interleukin 12 abrogate established experimental colitis in miceJ Exp Med19951825128112907595199
- SimpsonSJShahSComiskeyMT cell-mediated pathology in two models of experimental colitis depends predominantly on the interleukin 12/signal transducer and activator of transcription (Stat)-4 pathway, but is not conditional on interferon gamma expression by T cellsJ Exp Med19981878122512349547334
- BettelliECarrierYGaoWReciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cellsNature2006441709023523816648838
- BuonocoreSAhernPPUhligHHInnate lymphoid cells drive interleukin-23-dependent innate intestinal pathologyNature201046472931371137520393462
- AggarwalSGhilardiNXieMHde SauvageFJGurneyALInterleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17J Biol Chem200327831910191412417590
- StroberWZhangFKitaniAFussIFichtner-FeiglSProinflammatory cytokines underlying the inflammation of Crohn’s diseaseCurr Opin Gastroenterol201026431031720473158
- FussIJIs the Th1/Th2 paradigm of immune regulation applicable to IBD?Inflamm Bowel Dis200814Suppl 2S110S11218816734
- OlsenTRismoRCuiGGollRChristiansenIFlorholmenJTH1 and TH17 interactions in untreated inflamed mucosa of inflammatory bowel disease, and their potential to mediate the inflammationCytokine201156363364021945121
- KleinschekMABonifaceKSadekovaSCirculating and gut-resident human Th17 cells express CD161 and promote intestinal inflammationJ Exp Med2009206352553419273624
- RismoROlsenTCuiGNormalization of mucosal cytokine gene expression levels predicts long-term remission after discontinuation of anti-TNF therapy in Crohn’s diseaseScand J Gastroenterol201348331131923302000
- ChoiAMRyterSWLevineBAutophagy in human health and diseaseN Engl J Med2013368765166223406030
- MizushimaNYoshimoriTOhsumiYThe role of Atg proteins in autophagosome formationAnnu Rev Cell Dev Biol20112710713221801009
- MehrpourMEsclatineABeauICodognoPOverview of macroautophagy regulation in mammalian cellsCell Res201020774876220548331
- AlirezaeiMFoxHSFlynnCTElevated ATG5 expression in autoimmune demyelination and multiple sclerosisAutophagy20095215215819066443
- GukovskyIGukovskayaASImpaired autophagy underlies key pathological responses of acute pancreatitisAutophagy20106342842920215882
- MareninovaOAHermannKFrenchSWImpaired autophagic flux mediates acinar cell vacuole formation and trypsinogen activation in rodent models of acute pancreatitisJ Clin Invest2009119113340335519805911
- FujitaniYKawamoriRWatadaHThe role of autophagy in pancreatic beta-cell and diabetesAutophagy20095228028219158492
- IrelandJMUnanueERAutophagy in antigen-presenting cells results in presentation of citrullinated peptides to CD4 T cellsJ Exp Med2011208132625263222162830
- HanJWZhengHFCuiYGenome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosusNat Genet200941111234123719838193
- GatevaVSandlingJKHomGA large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosusNat Genet200941111228123319838195
- International Consortium for Systemic Lupus Erythemoatosus Genetics (SLEGEN)HarleyJBAlarcón-RiquelmeMEGenome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other lociNat Genet200840220421018204446
- PierdominiciMVomeroMBarbatiCRole of autophagy in immunity and autoimmunity, with a special focus on systemic lupus erythematosusFASEB J20122641400141222247332
- ChongSVickaryousNAsheAModifiers of epigenetic reprogramming show paternal effects in the mouseNat Genet200739561462217450140
- LevineBKroemerGAutophagy in the pathogenesis of diseaseCell20081321274218191218
- LevineBMizushimaNVirginHWAutophagy in immunity and inflammationNature2011469733032333521248839
- KuballaPNolteWMCastorenoABXavierRJAutophagy and the immune systemAnnu Rev Immunol20123061164622449030
- TravassosLHCarneiroLARamjeetMNod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entryNat Immunol2010111556219898471
- CooneyRBakerJBrainONOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentationNat Med2010161909719966812
- PlantingaTSCrisanTOOostingMCrohn’s disease-associated ATG16L1 polymorphism modulates pro-inflammatory cytokine responses selectively upon activation of NOD2Gut20116091229123521406388
- SanjuanMADillonCPTaitSWToll-like receptor signalling in macrophages links the autophagy pathway to phagocytosisNature200745071731253125718097414
- WaltzPCarchmanEHYoungACLipopolysaccaride induces autophagic signaling in macrophages via a TLR4, heme oxygenase-1 dependent pathwayAutophagy20117331532021307647
- SinghSBDavisASTaylorGADereticVHuman IRGM induces autophagy to eliminate intracellular mycobacteriaScience200631357921438144116888103
- DereticVMasterSSinghSAutophagy gives a nod and a wink to the inflammasome and Paneth cells in Crohn’s diseaseDev Cell200815564164219000829
- GutierrezMGMasterSSSinghSBTaylorGAColomboMIDereticVAutophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophagesCell2004119675376615607973
- McCarrollSAHuettAKuballaPDeletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s diseaseNat Genet20084091107111219165925
- LatianoAPalmieriOCucchiaraSPolymorphism of the IRGM gene might predispose to fistulizing behavior in Crohn’s diseaseAm J Gastroenterol2009104111011619098858
- SehgalRBergAPolinskiJIMutations in IRGM are associated with more frequent need for surgery in patients with ileocolonic Crohn’s diseaseDis Colon Rectum201255211512122228152
- BrestPLapaquettePSouidiMA synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s diseaseNat Genet201143324224521278745
- ManningGWhyteDBMartinezRHunterTSudarsanamSThe protein kinase complement of the human genomeScience200229856001912193412471243
- FrankeAMcGovernDPBarrettJCGenome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility lociNat Genet201042121118112521102463
- UmenoJAsanoKMatsushitaTMeta-analysis of published studies identified eight additional common susceptibility loci for Crohn’s disease and ulcerative colitisInflamm Bowel Dis201117122407241521351207
- BarrettJHIlesMMHarlandMGenoMEL ConsortiumGenome-wide association study identifies three new melanoma susceptibility lociNat Genet201143111108111321983787
- BerwickDCHarveyKLRRK2 signaling pathways: the key to unlocking neurodegeneration?Trends Cell Biol201121525726521306901
- Alegre-AbarrateguiJChristianHLufinoMMLRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular modelHum Mol Genet200918214022403419640926
- Gómez-SuagaPLuzón-ToroBChuramaniDLeucine-rich repeat kinase 2 regulates autophagy through a calcium-dependent pathway involving NAADPHum Mol Genet201221351152522012985
- MizushimaNThe role of the Atg1/ULK1 complex in autophagy regulationCurr Opin Cell Biol201022213213920056399
- HenckaertsLCleynenIBrinarMGenetic variation in the autophagy gene ULK1 and risk of Crohn’s diseaseInflamm Bowel Dis20111761392139721560199
- CleynenIGonzálezJRFigueroaCGenetic factors conferring an increased susceptibility to develop Crohn’s disease also influence disease phenotype: results from the IBDchip European ProjectGut Epub12212012
- Alvarez-LobosMArosteguiJISansMCrohn’s disease patients carrying Nod2/CARD15 gene variants have an increased and early need for first surgery due to stricturing disease and higher rate of surgical recurrenceAnn Surg2005242569370016244543
- RendaMCOrlandoACivitavecchiaGThe role of CARD15 mutations and smoking in the course of Crohn’s disease in a Mediterranean areaAm J Gastroenterol2008103364965518341489
- PragerMBüttnerJHaasVThe JAK2 variant rs10758669 in Crohn’s disease: altering the intestinal barrier as one mechanism of actionInt J Colorectal Dis201227556557322065112
- PierikMJoossensSVan SteenKToll-like receptor-1, -2, and -6 polymorphisms influence disease extension in inflammatory bowel diseasesInflamm Bowel Dis20061211816374251
- FarrellRJKelleherDGlucocorticoid resistance in inflammatory bowel diseaseJ Endocrinol2003178333934612967327
- PotocnikUFerkoljIGlavacDDeanMPolymorphisms in multidrug resistance 1 (MDR1) gene are associated with refractory Crohn disease and ulcerative colitisGenes Immun20045753053915505619
- CucchiaraSLatianoAPalmieriOItalian Society of Pediatric Gastroenterology and NutritionPolymorphisms of tumor necrosis factor-alpha but not MDR1 influence response to medical therapy in pediatric-onset inflammatory bowel diseaseJ Pediatr Gastroenterol Nutr200744217117917255827
- Van LimbergenJRussellRKDrummondHEDefinition of phenotypic characteristics of childhood-onset inflammatory bowel diseaseGastroenterology200813541114112218725221
- de RidderLWeersmaRKDijkstraGGenetic susceptibility has a more important role in pediatric-onset Crohn’s disease than in adult-onset Crohn’s diseaseInflamm Bowel Dis20071391083109217476680
- KugathasanSBaldassanoRNBradfieldJPLoci on 20q13 and 21q22 are associated with pediatric-onset inflammatory bowel diseaseNat Genet200840101211121518758464
- PetersonNGutherySDensonLGenetic variants in the autophagy pathway contribute to paediatric Crohn’s diseaseGut20085791336133718719149
- DubinskyMCWangDPicornellYet al; Western Regional Research Alliance for PediatricIBDIL-23 receptor (IL-23R) gene protects against pediatric Crohn’s diseaseInflamm Bowel Dis200713551151517309073
- Van LimbergenJRussellRKNimmoERIL23R Arg381Gln is associated with childhood onset inflammatory bowel disease in ScotlandGut20075681173117417337463
- ZhouZLinXYAkolkarPNVariation at NOD2/CARD15 in familial and sporadic cases of Crohn’s disease in the Ashkenazi Jewish populationAm J Gastroenterol200297123095310112492195
- VavassoriPBorgianiPBianconeLCARD15 mutation analysis in an Italian population: Leu1007fsinsC but neither Arg702Trp nor Gly908 Arg mutations are associated with Crohn’s diseaseInflamm Bowel Dis200410211612115168811
- ArnottIDNimmoERDrummondHENOD2/CARD15, TLR4 and CD14 mutations in Scottish and Irish Crohn’s disease patients: evidence for genetic heterogeneity within Europe?Genes Immun20045541742515190267
- GazouliMMantzarisGKotsinasAAssociation between polymorphisms in the Toll-like receptor 4, CD14, and CARD15/NOD2 and inflammatory bowel disease in the Greek populationWorld J Gastroenterol200511568168515655821
- BaldassanoRNBradfieldJPMonosDSAssociation of the T300A non-synonymous variant of the ATG16L1 gene with susceptibility to paediatric Crohn’s diseaseGut20075681171117317625155
- PrescottNJDominyKMKuboMIndependent and population-specific association of risk variants at the IRGM locus with Crohn’s diseaseHum Mol Genet20101991828183920106866
- RiouxJDDalyMJSilverbergMSGenetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn diseaseNat Genet200129222322811586304
- SilverbergMSDuerrRHBrantSRNIDDK IBD Genetics ConsortiumRefined genomic localization and ethnic differences observed for the IBD5 association with Crohn’s diseaseEur J Hum Genet200715332833517213842
- TomerGWetzlerGKeddacheMDensonLAPolymorphisms in the IBD5 locus are associated with Crohn disease in pediatric Ashkenazi Jewish patientsJ Pediatr Gastroenterol Nutr200948553153719412005
- Diaz-GalloLMMedranoLMGómez-GarcíaMAnalysis of the influence of two CD24 genetic variants in Crohn’s disease and ulcerative colitisHum Immunol2011721096997221684315
- BarrettJCHansoulSNicolaeDLGenome-wide association defines more than 30 distinct susceptibility loci for Crohn’s diseaseNat Genet200840895596218587394
- ZhuHLiYROxidative stress and redox signaling mechanisms of inflammatory bowel disease: updated experimental and clinical evidenceExp Biol Med (Maywood)2012237547448022442342
- AtanDHeissigerovaJKuffováLTumor necrosis factor polymorphisms associated with tumor necrosis factor production influence the risk of idiopathic intermediate uveitisMol Vis20131918419523378732
- AldhousMCNimmoERSatsangiJNOD2/CARD15 and the Paneth cell: another piece in the genetic jigsaw of inflammatory bowel diseaseGut200352111533153514570717
- IwanagaYDaveyMPMartinTMCloning, sequencing and expression analysis of the mouse NOD2/CARD15 geneInflamm Res200352627227612835899
- RockFLHardimanGTimansJCKasteleinRABazanJFA family of human receptors structurally related to Drosophila TollProc Natl Acad Sci USA19989525885939435236
- ParhamCChiricaMTimansJA receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23RJ Immunol2002168115699570812023369
- GründemannDHarlfingerSGolzSDiscovery of the ergothioneine transporterProc Natl Acad Sci USA2005102145256526115795384
- ZhengHJiCLiJCloning and analysis of human Apg16LDNA Seq200415430330515620219
- BekpenCXavierRJEichlerEEHuman IRGM gene “to be or not to be”Semin Immunopathol201032443744420737271
- FriedrichsFStollMRole of discs large homolog 5World J Gastroenterol200612233651365616773680
- OrensteinSJKuoSHTassetIInterplay of LRRK2 with chaperone-mediated autophagyNat Neurosci201316439440623455607
- VangTMileticAVBottiniNMustelinTProtein tyrosine phosphatase PTPN22 in human autoimmunityAutoimmunity200740645346117729039
- LiuYWeiSHHoASde Waal MalefytRMooreKWExpression cloning and characterization of a human IL-10 receptorJ Immunol19941524182118298120391
- LocksleyRMKilleenNLenardoMJThe TNF and TNF receptor superfamilies: integrating mammalian biologyCell2001104448750111239407