Abstract
Mitochondrial DNA mutations play an important role in causing sensorineural hearing loss. The purpose of this study was to determine the association of the mitochondrial genes RNR1, MT-TL1, and ND1 as well as the nuclear genes GJB2 and GJB6 with audiological examinations in nonfamilial Iranians with cochlear implants, using polymerase chain reaction, DNA sequencing, and RNA secondary structure analysis. We found that there were no novel mutations in the mitochondrial gene 12S rRNA (MT-RNR1) in patients with and without GJB2 mutation (GJB2+ and GJB2−, respectively), but a total of six polymorphisms were found. No mutations were observed in tRNALeu(UUR) (MT-TL1). Furthermore, eight polymorphisms were found in the mitochondrial ND1 gene. Additionally, no mutations were observed in the nuclear GJB6 gene in patients in the GJB2− and GJB2+ groups. The speech intelligibility rating and category of auditory perception tests were statistically assessed in patients in the GJB2− and GJB2+ groups. The results indicated that there was a significant difference (P<0.05) between the categories of auditory perception score in the GJB2− group compared to that in the GJB2+ group. Successful cochlear implantation was observed among individuals with GJB2 mutations (GJB2+) and mitochondrial polymorphisms compared to those without GJB2 mutations (GJB2−). In conclusion, the outcome of this study suggests that variation in the mitochondrial and nuclear genes may influence the penetrance of deafness. Therefore, further genetic and functional studies are required to help patients in making the best choice for cochlear implants.
Introduction
Hearing loss (HL) is one of the most common sensory disorders in humans affecting one to three of every 1,000 newborns.Citation1 The onset of HL usually occurs in childhood, is predominantly postlingual, and may be accompanied by vertigoCitation2 and tinnitus.Citation3,Citation4 There is a high variability in severity, ranging from normal hearing to profound deafness.Citation5,Citation6 This may be due to the fact that the phenotypic effects are a result of several factors and can develop gradually.Citation7 HL occurs in both syndromic and nonsyndromic deafness caused by mitochondrial DNA (mtDNA) mutations,Citation8 where both environmental and genetic factors are also involved, such as noise pollution, use of aminoglycoside drugs, and genomic diversity.Citation9 mtDNA mutations are responsible for both maternally inherited syndromic and nonsyndromic HL and play a role in predisposition to aminoglycoside-induced ototoxicity.Citation10 In Italy, at least 5% of cases of postlingual, non-syndromic hearing impairment may be attributed to mtDNA mutations.Citation1 Furthermore, it has been estimated that in up to 67% of patients with and without GJB2 mutations (GJB2+ and GJB2−, respectively), mtDNA disorders also manifest as sensorineural hearing loss (SNHL).Citation11 SNHL associated with mtDNA mutations is generally progressive with high frequency.Citation12–Citation15 This may be explained by the high oxidative phosphorylation demands in cochlear cells, as conveyed by mtDNA mutations.Citation1
Human mtDNA is a 16,569-bp, circular, double-stranded molecule that encodes 37 genes, including 13 subunits of the respiratory chain complexes, two ribosomal RNAs, and 22 transfer RNAs. Each nucleated human cell contains a few thousand copies of mtDNA. The somatic mutation rate of mtDNA is presumed to be 10–20 times higher than that of nuclear DNA (nDNA).Citation16 Mitochondria are essentially double-membraned subcellular organelles present in all nucleated mammalian cells. Their primary function is to support aerobic respiration, that is, the production of adenosine triphosphate through oxidative phosphorylation.Citation17 In addition, mutations and/or polymorphism variance in mitochondrial genes play important roles and are related to many diseases, such as Leber’s hereditary optic neuropathy,Citation18 Friedreich’s ataxia,Citation19 autism,Citation20 Alzheimer’s disease,Citation21 oculocutaneous albinism type 1,Citation22 recurrent pregnancy loss,Citation23 and different cancers, such as gastric,Citation24,Citation25 bladder,Citation26 colorectal,Citation27 and breast.Citation24
HL is caused by genetic or nongenetic factors. The nongenetic risk factors for HL during the neonatal period include treatment in a neonatal intensive care unit, craniofacial anomalies, meningitis,Citation28,Citation29 and cytomegalovirus infections.Citation30 mtDNA variants, including mutations, deletions, and insertions, particularly in the MT-RNR1 gene, have been identified to play an important role in patients with SNHL associated with or without a history of aminoglycoside therapy, suggesting that this locus in particular is a hotspot for deafness-associated mutations.Citation31,Citation32 The MT-TL1 gene encoding mitochondrial tRNALeu(UUR) is a hotspot for pathogenic mtDNA mutations,Citation33 and a previous study reported the possible role of MT-TL1 in nonsyndromic disease.Citation9 Mutations in this region cause severe myopathy with respiratory insufficiency,Citation33,Citation34 as this region is highly conserved among mammals.Citation35 mtDNA mutations, in particular T3308C, have been identified to induce a significant decrease in the levels of the MT-ND1 gene, suggesting that mutations in this region can increase the penetrance of deafness in patients with HL.Citation1,Citation36 Mutations in the nuclear GJB2 and GJB6 genes on the DFNB1 locus at chromosome 13q11-q12 are responsible for up to 50% of the most common causes of prelingual onset, recessively inherited nonsyndromic SNHL in humans, encoding the gap junction proteins connexin 26 (Cx26) and connexin 30, respectively,Citation37,Citation38 and play a role in cochlear homeostasis.Citation39 Recessive mutations in the GJB2 gene are the most common cause of hearing impairmentCitation40 affecting both paternal and maternal alleles.Citation40,Citation41 Thus, in order to estimate the incidence ratio of mutation in the next generation, the frequency of the mutation is to be ascertained.Citation41,Citation42 The GJB2 gene is the most common cause of the congenital HL,Citation43 and the mutation spectra are different among different ethnic groups.Citation42 It is essential to investigate the carrier frequency and mutation spectrum of each genetic background in order to provide more precise genetic counseling. The GJB2 gene encodes Cx26, a member of the connexin family of proteins that are constituents of the intercellular gap junction.Citation44 The first GJB2 mutation was reported by Kelsell et al,Citation45 in which c.35delG is the most common mutation in the Caucasian population while c.235delC and p.Val37Ile are the most common mutations in the Asian population.Citation42,Citation46 In this study, we aimed to compare the impact of mutations in the mitochondrial 12S rRNA (MT-RNR1), tRNALeu(UUR) (MT-TL1), and ND1 on Iranian patients with nuclear GJB2 mutation (GJB2+) and without GJB2 mutation (GJB2−) undergoing cochlear implants.
Materials and methods
Specimen collection and ethical statement
In this study, blood samples from 84 patients who had prelingual deafness with normal cochlear structures were obtained from the Rasoul Akram Hospital, Tehran, Iran. The patients did not show any syndromic symptoms or other clinical abnormalities, including muscular diseases, diabetes, visual dysfunction, or neurological disorders. Eighty-four patients with cochlear implant were categorized into two groups including 24 patients without GJB2 (GJB2−) mutations and 60 patients with GJB2 (GJB2+) mutations. Written informed consent, including consent to participate in this study and consent to submission and publish, was obtained from the parents on behalf of their children in accordance with the Medical Ethics Committee of Rasoul Akram Hospital, Tehran, Iran (Approval No 375/105/D/93).
Audiological examinations
Categories of auditory performance (CAP)Citation47 and speech intelligibility rating (SIR)Citation48 were used to assess hearing ability and speech intelligibility, respectively. The infants were screened at 2 months of age and were then referred to an audiologist after 2 years of age for audiometric tests, including CAP and SIR tests, where profound HLs (>90 dB) were identified in the patients. In the CAP test, the rating of auditory ability consists of the following eight categories:Citation47 Score 0 – no awareness of environmental sounds; Score 1 – awareness of environmental sounds; Score 2 – response to speech sounds; Score 3 – recognition of environmental sounds; Score 4 – discrimination of at least two speech sounds; Score 5 – understanding of common phrases without lip reading; Score 6 – understanding of conversation without lip reading with a familiar talker; and Score 7 – use of a telephone with a familiar talker. The SIR test consists of five categories.Citation48 Scores 1–5 depend on the spoken ability rating scale criteria, which are as follows: 5 – connected speech is intelligible to all listeners and the child is understood easily in everyday contexts; 4 – connected speech is intelligible to a listener who has a little experience of a deaf person’s speech; 3 – connected speech is intelligible to a listener who concentrates and lip-reads; 2 – connected speech is unintelligible, where intelligible speech is developing single words when context and lip-reading cues are available; and 1 – connected speech is unintelligible, where prerecognizable words in spoken language, the primary mode of communication, may be manual. All children received early intervention services within an average of 2 months after identification. This study was conducted in the Cochlear Implant Centre of the Hazrat Rasoul Akram Medical Complex, Tehran, Iran. The CAP and SIR assessments were conducted by two speech therapists during the follow-up period before implantation and at 6 months, 1 year, and 2 years after implantation. All 84 subjects had scores for both CAP and SIR before and after implantation. The children had to orally repeat the words/sentences they heard from the test conductor who produced the stimuli with his/her mouth covered (all the subjects were tested by the same conductor) and were scored based on the number of (key)words they correctly repeated. The answers were then recorded.
DNAextraction and polymerase chain reaction
Total DNA was extracted from the peripheral blood sample of each individual using a QIAmap DNA Micro Kit (QIAGEN no 56304). The DNA was amplified for the mitochondrial 12S rRNA, tRNALeu(UUR), and ND1 genes (located near the MT-TL1 gene) and also for the nuclear GJB2 and GJB6 genes, using specific primers ().
Table 1 PCR primers of selected nuclear and mitochondrial genes
Briefly, polymerase chain reaction (PCR) was performed in 25 μL of reaction volumes containing 50–100 ng of genomic DNA, 2.5 μL of 10× PCR buffer, 10 mM of each dNTP, 1 mM of MgCl2, 10 pmol of each primer, and 5 U of Taq polymerase (CinnaGen, Tehran, Iran) to a final volume of 25 μL, topped up using distilled water. The reactions were performed in a thermal cycler (Eppendorf, Humburg, Germany), and the PCR products were examined on 1.5% agarose gel electrophoresis. The PCR conditions for amplification of the mitochondrial 12S rRNA were as follows: initial DNA denaturation at 95°C for 5 minutes, then 35 cycles of denaturation at 95°C for 1 minute, annealing at 50°C for 1 minute, and extension at 72°C for 1 minute, followed by final extension at 72°C for 10 minutes. The PCR conditions for amplification of the mitochondrial tRNALeu(UUR) were as follows: initial DNA denaturation at 94°C for 5 minutes, then 35 cycles of denaturation at 94°C for 50 seconds, annealing at 55°C for 50 seconds, and extension at 72°C for 45 seconds, followed by final extension at 72°C for 10 minutes.
The primer sequences for the GJB6 gene used in this study were in accordance with previous studies.Citation49–Citation51 How ever, neither mutations nor deletions have been identified in the GJB6 gene in our patients. The PCR conditions for amplification of the nuclear GJB6 gene were as follows: initial DNA denaturation at 95°C for 5 minutes, then 35 cycles of denaturation at 95°C for 1 minute, annealing at 58.5°C for 1 minute, and extension at 72°C for 1 minute, followed by final extension at 72°C for 10 minutes. The PCR conditions for amplification of the nuclear GJB2 gene were performed in two steps, after initial DNA denaturation at 93°C for 3 minutes. The first step was done with five cycles of denaturation at 95°C for 1 minute, annealing at 59°C for 1 minute, and then final extension at 72°C for 1 minute. The second step was carried out with 26 cycles of denaturation at 94°C for 45 seconds, annealing at 59°C for 45 seconds, and extension at 72°C for 45 seconds, followed by final extension at 72°C for 8 minutes.
DNA sequencing and analysis of variants
The PCR products were sequenced with the respective forward or reversed primers on an ABI 3700 sequencer (Takapo Zist Company, Tehran, Iran) and compared with the revised Cambridge Reference Sequence and the NCBI Reference Sequence Database (http://www.ncbi.nlm.nih.gov/refseq/) using the FinchTV program version 1.4.0. Identification of the nucleotide changes was verified through MITOMAP (http://mitomap.org/MITOMAP) and the Human Gene Mutation Database, 2007.
RNA secondary structure analysis
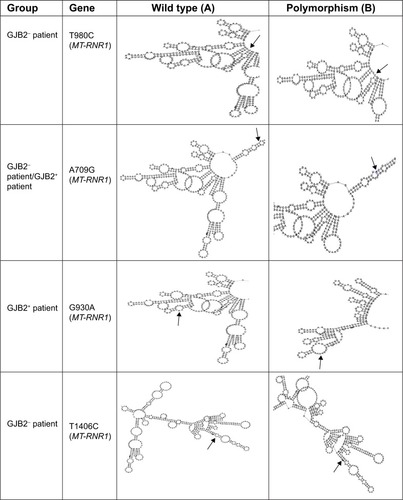
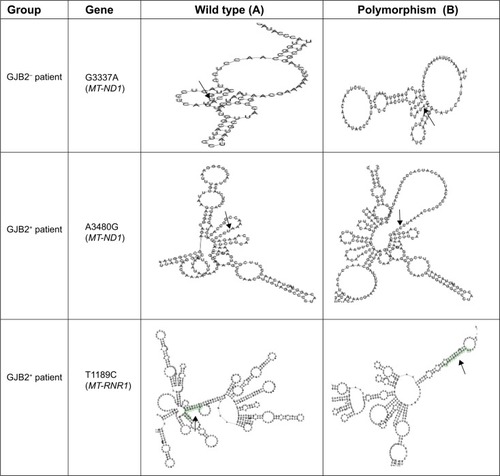
The RNA fold software from the Institute for Theoretical Chemistry, University of Vienna ([email protected]; http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi), was used to predict the RNA secondary structure based on minimum energy requirements and base pairing ( and ).
Figure 1 Comparison of sequence analysis of the polymorphism in MT-RNR1 and MT-ND1 and heterozygous GJB2 genes among patients with GJB2 mutation (GJB2+) and without GJB2 mutation (GJB2−).
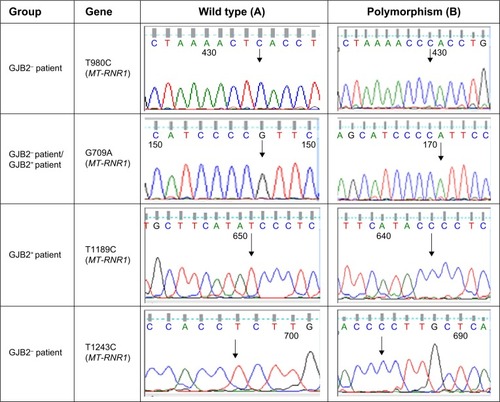
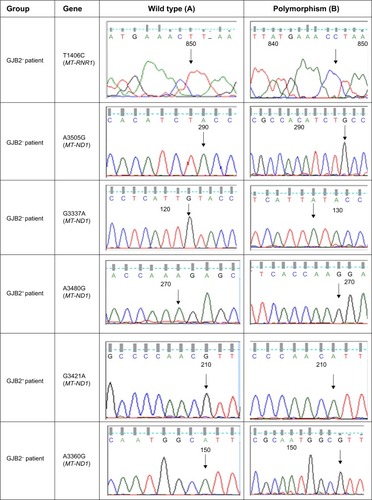
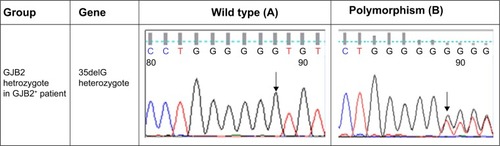
Figure 2 Comparison of RNA secondary structure analysis of wild type and polymorphic predicted in MT-RNR1 and MT-ND1 genes among patients with GJB2 mutation (GJB2+) and without GJB2 mutation (GJB2−).
Statistical analysis
The independent t-test using Statistical Package for the Social Sciences, version 13, was used to analyze the relationship between CAP and SIR with cochlear implantation in the GJB2− and GJB2+ groups; P-values <0.05 were regarded as statistically significant ().
Table 2 Statistical analysis of SIR and CAP in the GJB2− and GJB2+ patients using independent t-test
Results
In this study, the MT-RNR1 (12sRNA) and MT-TL1 (tRNALeu(UUR)) genes were analyzed. Polymorphisms that were detected in MT-RNR1 included G709A in eight patients (9.6%), T1243C in two patients (2.4%), T980C in three patients (3.6%), T1406C in one patient (1.2%), G930A in one patient (1.2%), and T1189C in three patients (3.6%). No mutation was found in the MT-TL1 (tRNALeu(UUR)) gene. Additionally, eight polymorphisms in the MT-ND1 (NADH dehydrogenase I) gene were found, including A3360G (1.2%), A3505G (2.4%), A3339G (1.2%), G3337A (2.4%), G3392C (1.2%), G3483A (1.2%), A3480G (3.6%), and G3421A (2.4%). Here, all mtDNA mutations were homoplasmic, while only one – A3339G (1.2%) – was heteroplasmic mtDNA ND1 polymorphism. Two GJB2+ patients with V153I and R184P did not show any mitochondrial RNR1 and ND1 mutations. In addition, all GJB2− and GJB2+ patients revealed the presence of the A750G and A1438G polymorphisms in the MT-RNR1 gene. Sequence analysis in the nuclear GJB2 and GJB6 genes demonstrated that no mutation in nuclear GJB6 gene was detected in all GJB2− and GJB2+ groups. Of the 84 individuals, no GJB2 mutation was observed in 24 patients (GJB2−), but different types of GJB2 mutations were observed among the 60 GJB2+ patients (; ).
Figure 3 Frequency of polymorphism MT-RNR1 and MT-ND1 genes in patients without GJB2 mutation (GJB2−) and patients with GJB2 mutation (GJB2+).
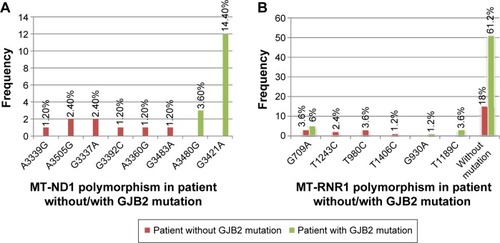
Table 3 Summary of clinical data and mitochondrial RNR1 and ND1 and nuclear GJB2 and GJB6 gene alterations
Discussion
In this study, the impact of the mitochondrial 12S rRNA (MT-RNR1), tRNALeu(UUR) (MT-TL1), and ND1 variations among 84 random Iranian patients were compared between patients with GJB2 mutation (GJB2+) and without GJB2 mutation (GJB2−) undergoing cochlear implants, which had not been examined by earlier Iranian researchers. These hotspots are recognized to be among the most frequent causes of hearing impairment.Citation52 Mitochondria play an important function in metabolism, thus mutations in mitochondrial genes result in many metabolic diseases.Citation53 In the present study, we analyzed three fragments of mtDNA, namely 12S rRNA (MT-RNR1), tRNALeu(UUR) (MT-TL1), and ND1, and two fragments of nDNA, namely GJB2 (Cx26) and GJB6, in 84 subjects with nonsyndromic HL. All patients (GJB2− and GJB2+) were found to have the A750G and A1438G polymorphisms in the MT-RNR1 gene, which is in agreement with previous studies that reported that the A750G, A1438G, A4769G, A8860G, and A15326G polymorphisms are common to mtDNA sequences from Africans, Asians, and Europeans.Citation54 Therefore, the polymorphisms in the MT-RNR1 gene are not the cause of deafness and hearing impairment in Iranian patients. However, among the eight polymorphisms in the MT-ND1 (NADH dehydrogenase I) gene, A3505G and A3480G are associated with prostate cancer, while G3337A and G3421A are associated with cardiomyopathyCitation55 and pancreatic cancer,Citation56 respectively.
It should be noted that late-onset and gradual worsening of hearing impairment may reflect the tendency of the mitochondrion to accumulate mutations with aging, due to its genomic instability. As previously reported,Citation57 mitochondrial mutation plays an important role in cochlear implantation, whereas in our study, mitochondrial polymorphisms also indicate candidacy for cochlear implantation. Additionally, RNA secondary structure analysis demonstrated that the RNA at G930A, T1189C, and A3480G had different structures to compare the wild type, where it is possible that these polymorphisms and their correlation with the GJB2 mutation have more effect on deafness.
The association between mitochondrial and nuclear mutations with aminoglycoside exposure is quite variable, where aminoglycosides are probably only one of the factors interacting with the mutation in determining the deafness phenotype and play a role only in ~20% of patients.Citation1,Citation58–Citation61 Several studies have reported that patients carrying the A1555G mutation exposed to aminoglycosides develop more severe deafness with an earlier onset.Citation31,Citation59,Citation60,Citation62–Citation64 However, consistent with the other studies,Citation58–Citation61,Citation65,Citation66 our study showed that even in the absence of aminoglycoside exposure, mitochondrial polymorphisms could be responsible for hearing impairment. Previous studies revealed the mitochondrial m1555A>G,Citation67 m.1005T>C,Citation68 and m.921T.CCitation68 mutations among Iranian patients, while these mentioned mutations were not detected in our patients with HL. Additionally, a study indicated the high carrier frequency of the nuclear GJB2 mutation (35delG) in the north of Iran.Citation69
Conclusion
In conclusion, our finding indicates that a combination of polymorphisms in mitochondrial and nuclear genes may increase the penetrance of deafness and that there may be an association between nuclear gene variation and mitochondrial deafness with cochlear implant in Iranian patients. Therefore, the pathogenicity impact of mtDNA and nDNA variants and their correlation with other conditions should be established. Therefore, further genetic and functional studies are required in order to help individuals decide whether to undergo cochlear implantation.
Author contributions
Conceived and designed the experiments: MH. Performed the experiments: MB. Analyzed the data and contributed the reagents/materials/analysis tools: MDA, MRH, HA, and MF. Wrote the manuscript, contributed to the discussion, and reviewed the article: BK and FA. All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Acknowledgments
We would like to thank all the participants from the Rasoul Akram Hospital, Tehran, Iran, for blood donation.
Disclosure
The authors report no conflicts of interest in this work.
References
- GuaranVAstolfiLCastiglioneAAssociation between idiopathic hearing loss and mitochondrial DNA mutations: a study on 169 hearing-impaired subjectsInt J Mol Med201332478579423969527
- ChenJNHoKYJuanKHSensorineural hearing loss in MELAS syndrome – case reportKaohsiung J Med Sci19981485195239780603
- YuanHJiangSYangWGuoWCaoJDaiP[Screening for mitochondrial 1555(G) mutation in patients with aminoglycoside antibiotic-induced deafness]Zhonghua Yi Xue Yi Chuan Xue Za Zhi199916314114410359861
- SimdonJWattersDBartlettSConnickEOtotoxicity associated with use of nucleoside analog reverse transcriptase inhibitors: a report of 3 possible cases and review of the literatureClin Infect Dis200132111623162711340535
- BravoOBallanaEEstivillXCochlear alterations in deaf and unaffected subjects carrying the deafness-associated A1555G mutation in the mitochondrial 12S rRNA geneBiochem Biophys Res Commun2006344251151616631122
- YoungWYZhaoLQianYVariants in mitochondrial tRNAGlu, tRNAArg, and tRNAThr may influence the phenotypic manifestation of deafness-associated 12S rRNA A1555G mutation in three Han Chinese families with hearing lossAm J Med Genet A2006140202188219716955413
- ZhaoHLiRWangQMaternally inherited aminoglycoside-induced and nonsyndromic deafness is associated with the novel C1494T mutation in the mitochondrial 12S rRNA gene in a large Chinese familyAm J Hum Genet200474113915214681830
- XingGChenZCaoXMitochondrial rRNA and tRNA and hearing functionCell Res200717322723917199108
- BerrettiniSForliFPassettiSMitochondrial non-syndromic sensorineural hearing loss: a clinical, audiological and pathological study from Italy, and revision of the literatureBiosci Rep2008281495918215147
- MiyagawaMNishioSYIkedaTFukushimaKUsamiSIMassively parallel DNA sequencing successfully identifies new causative mutations in deafness genes in patients with cochlear implantation and EASPLoS One2013810e7579324130743
- LiJZHuYQWangSH[Mutations of Cx26 gene in patients with NSHL and intracellular distribution of two mutants]Yi Chuan200931770571219586875
- LiZLiRChenJMutational analysis of the mitochondrial 12S rRNA gene in Chinese pediatric subjects with aminoglycoside-induced and non-syndromic hearing lossHum Genet2005117191515841390
- BallanaEMoralesERabionetRMitochondrial 12S rRNA gene mutations affect RNA secondary structure and lead to variable penetrance in hearing impairmentBiochem Biophys Res Commun2006341495095716458854
- GurtlerNSchmuzigerNKimYMhatreANJungiMLalwaniAKAudiologic testing and molecular analysis of 12S rRNA in patients receiving aminoglycosidesLaryngoscope2005115464064415805873
- ScagliaFHsuCHKwonHMolecular bases of hearing loss in multi-systemic mitochondrial cytopathyGenet Med200681064165217079881
- ShenkarRNavidiWTavaréSThe mutation rate of the human mtDNA deletion mtDNA4977Am J Hum Genet19965947728808591
- HatefiYThe mitochondrial electron transport and oxidative phosphorylation systemAnnu Rev Biochem1985541101510692862839
- RezvaniZDidariEArastehkaniAFifteen novel mutations in the mitochondrial NADH dehydrogenase subunit 1, 2, 3, 4, 4L, 5 and 6 genes from Iranian patients with Leber’s hereditary optic neuropathy (LHON)Mol Biol Rep201340126837684124158608
- SalehiMHKamalidehghanBHoushmandMGene expression profiling of mitochondrial oxidative phosphorylation (OXPHOS) complex I in Friedreich ataxia (FRDA) patientsPLoS One201494e9406924705504
- MousavizadehKAskariMArianHAssociation of human mtDNA mutations with autism in Iranian patientsJ Res Med Sci2013181092624497871
- NiaSSAzadfarPAkbariLNew pathogenic variations of mitochondrial DNA in Alzheimer disease!J Res Med Sci201318326926923930130
- KalahroudiVGKamalidehghanBKaniAATwo novel tyrosinase (TYR) gene mutations with pathogenic impact on oculocutaneous albinism type 1 (OCA1)PLoS One201499e10665625216246
- ColagarAHMosaiebyESeyedhassaniSMT4216C mutation in NADH dehydrogenase I gene is associated with recurrent pregnancy lossMitochondrial DNA201324561061223464625
- GhaffarpourMMahdianRFereidooniFKamalidehghanBMoazamiNHoushmandMThe mitochondrial ATPase6 gene is more susceptible to mutation than the ATPase8 gene in breast cancer patientsCancer Cell Int2014142124588805
- AndersonSBankierATBarrellBGSequence and organization of the human mitochondrial genomeNature198129058064574657219534
- ShakhssalimNHoushmandMKamalidehghanBThe mitochondrial C16069T polymorphism, not mitochondrial D310 (D-loop) mononucleotide sequence variations, is associated with bladder cancerCancer Cell Int201313112024308421
- AkouchekianMHoushmandMAkbariMHHKamalidehghanBDehghanMAnalysis of mitochondrial ND1 gene in human colorectal cancerJ Res Med Sci20111615021448383
- SuttonGJRoweSJRisk factors for childhood sensorineural hearing loss in the Oxford regionBr J Audiol199731139549056042
- PediatricsAAOAssociation AS-L-HYear 2000 position statement: principles and guidelines for early hearing detection and intervention programsPediatrics2000106479881711015525
- NanceWELimBGDodsonKMImportance of congenital cytomegalovirus infections as a cause for pre-lingual hearing lossJ Clin Virol200635222122516384744
- CasanoRAJohnsonDFBykhovskayaYTorricelliFBigozziMFischel-GhodsianNInherited susceptibility to aminoglycoside ototoxicity: genetic heterogeneity and clinical implicationsAm J Otolaryngol199920315115610326749
- TangH-YHutchesonENeillSDrummond-BorgMSpeerMAlfordRLGenetic susceptibility to aminoglycoside ototoxicity: how many are at risk?Genet Med20024533634512394346
- BindoffLHowellNPoultonJAbnormal RNA processing associated with a novel tRNA mutation in mitochondrial DNA. A potential disease mechanismJ Biol Chem19932682619559195648366098
- van den BoschBde CooIHendrickxAIncreased risk for cardiorespiratory failure associated with the A3302G mutation in the mitochondrial DNA encoded tRNALeu (UUR) geneNeuromuscul Disord2004141068368815351426
- HelmMBruléHFriedeDGiegéRPützDFlorentzCSearch for characteristic structural features of mammalian mitochondrial tRNAsRNA20006101356137911073213
- LiXFischel-GhodsianNSchwartzFYanQFriedmanRAGuanMXBiochemical characterization of the mitochondrial tRNASer (UCN) T7511C mutation associated with nonsyndromic deafnessNucleic Acids Res200432386787714960712
- WilchEZhuMBurkhartKBExpression of GJB2 and GJB6 is reduced in a novel DFNB1 alleleAm J Hum Genet200679117417916773579
- GrifaAWagnerCAD’AmbrosioLMutations in GJB6 cause nonsyndromic autosomal dominant deafness at DFNA3 locusNat Genet1999231161810471490
- ZhaoH-BKikuchiTNgezahayoAWhiteTGap junctions and cochlear homeostasisJ Membr Biol20062092–317718616773501
- TaniguchiMMatsuoHShimizuSCarrier frequency of the GJB2 mutations that cause hereditary hearing loss in the Japanese populationJ Hum Genet2015601061361726178431
- AbeSUsamiSShinkawaHKelleyPMKimberlingWJPrevalent connexin 26 gene (GJB2) mutations in JapaneseJ Med Genet2000371414310633133
- OhtsukaAYugeIKimuraSGJB2 deafness gene shows a specific spectrum of mutations in Japan, including a frequent founder mutationHum Genet2003112432933312560944
- SmithRJBaleJFWhiteKRSensorineural hearing loss in childrenLancet2005365946287989015752533
- KumarNMGilulaNBThe gap junction communication channelCell19968433813888608591
- KelsellDDunlopJStevensHConnexin 26 mutations in hereditary non-syndromic sensorineural deafnessNature1997387662880839139825
- GreenGEScottDAMcDonaldJMWoodworthGGSheffieldVCSmithRJCarrier rates in the midwestern United States for GJB2 mutations causing inherited deafnessJAMA1999281232211221610376574
- ArchboldSLutmanMEMarshallDHCategories of auditory performanceAnn Otol Rhinol Laryngol Suppl19951663123147668685
- AllenCNikolopoulosTPDyarDO’DonoghueGMReliability of a rating scale for measuring speech intelligibility after pediatric cochlear implantationOtol Neurotol200122563163311568670
- KahriziKShafeghatiYDaneshiAJogataieM-TDelta (GJB6-D13S1830) is not a common cause of nonsyndromic hearing loss in the Iranian populationArch Iran Med200582104108
- MahdiehNRaeisiMShirkavandABagherianHAkbariMTZeinaliSInvestigation of GJB6 large deletions in Iranian patients using quantitative real-time PCRClin Lab2009569–1046747121086793
- SadeghiASanatiMHAlastiFHashemzadeh ChaleshtoriMAtaeiMMutation analysis of connexin 26 gene and del (GJB6-D13S1830) in patients with hereditary deafness from two provinces in IranIran J Biotechnol20053255258
- DzhemilevaLUPosukhOLTazetdinovAM[Analysis of mitochondrial 12S rRNA and tRNA(Ser(UCN)) genes in patients with nonsyndromic sensorineural hearing loss from various regions of Russia]Genetika200945798299119705751
- ShadelGSExpression and maintenance of mitochondrial DNA: new insights into human disease pathologyAm J Pathol200817261445145618458094
- HerrnstadtCElsonJLFahyEReduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroupsAm J Hum Genet20027051152117111938495
- BrandonMBaldiPWallaceDCMitochondrial mutations in cancerOncogene200625344647466216892079
- JansenRJFonseca-WilliamsSBamletWRDetection of DNA damage in peripheral blood mononuclear cells from pancreatic cancer patientsMol Carcinog201554101220122625111947
- SudoATakeichiNHosokiKSaitohSSuccessful cochlear implantation in a patient with mitochondrial hearing loss and m.625G>A transitionJ Laryngol Otol2011125121282128521914246
- Fischel-GhodsianNMitochondrial deafnessEar Hear200324430331312923421
- CasanoRABykhovskayaYJohnsonDFHearing loss due to the mitochondrial A1555G mutation in Italian familiesAm J Med Genet19987953883919779807
- EstivillXGoveaNBarceloAFamilial progressive sensorineural deafness is mainly due to the mtDNA A1555G mutation and is enhanced by treatment with aminoglycosidesAm J Human Genet199862127359490575
- PrezantTRAgapianJVBohlmanMCMitochondrial ribosomal RNA mutation associated with both antibiotic–induced and non–syndromic deafnessNat Genet1993432892947689389
- BravermanIJaberLLeviHAudiovestibular findings in patients with deafness caused by a mitochondrial susceptibility mutation and precipitated by an inherited nuclear mutation or aminoglycosidesArch Otolaryngol Head Neck Surg19961229100110048797567
- El-SchahawiMde MunainALSarrazinATwo large Spanish pedigrees with nonsyndromic sensorineural deafness and the mtDNA mutation at nt 1555 in the 12S rRNA gene evidence of heteroplasmyNeurology19974824534569040738
- NoguchiYYashimaTItoTSumiTTsuzukuTKitamuraKAudio-vestibular findings in patients with mitochondrial A1555G mutationLaryngoscope2004114234434814755216
- HutchinTHigashiKFischel-GhodsianNStonekingMArnosCA molecular basis for human hypersensitivity of aminoglyscoside antibioticsNucleic Acids Res19932118417441798414970
- MatsunagaTKumanomidoHShiromaMOhtsukaAAsamuraKUsamiSiDeafness due to A1555G mitochondrial mutation without use of aminoglycosideLaryngoscope200411461085109115179218
- ZohourMMAkbariMChaleshtoriMHFrequency of the mitochondrial A1555G mutation in Iranian patients with non-syndromic hearing impairmentIndian J Sci Technol201251033783383
- DowlatiMADerakhshandeh-peykarPHoushmandMNovel nucleotide changes in mutational analysis of mitochondrial 12SrRNA gene in patients with nonsyndromic and aminoglycoside-induced hearing lossMol Biol Rep20134032689269523242658
- ChaleshtoriMHFarrokhiEShahraniMHigh carrier frequency of the GJB2 mutation (35delG) in the north of IranInt J Pediatr Otorhinolaryngol200771686386717428550
