Abstract
Prion diseases are fatal neurodegenerative disorders that affect animals and humans. There is a need to gain understanding of prion disease pathogenesis and to develop diagnostic assays to detect prion diseases prior to the onset of clinical symptoms. The goal of this study was to identify genes that show altered expression early in the disease process in the spleen and brain of prion disease-infected mice. Using Affymetrix microarrays, we identified 67 genes that showed increased expression in the brains of prion disease-infected mice prior to the onset of clinical symptoms. These genes function in many cellular processes including immunity, the endosome/lysosome system, hormone activity, and the cytoskeleton. We confirmed a subset of these gene expression alterations using other methods and determined the time course in which these changes occur. We also identified 14 genes showing altered expression prior to the onset of clinical symptoms in spleens of prion disease infected mice. Interestingly, four genes, Atp1b1, Gh, Anp32a, and Grn, were altered at the very early time of 46 days post-infection. These gene expression alterations provide insights into the molecular mechanisms underlying prion disease pathogenesis and may serve as surrogate markers for the early detection and diagnosis of prion disease.
Keywords:
Background
Transmissible spongiform encephalopathies (TSEs), also termed prion diseases, are fatal neurodegenerative disorders that occur in humans and various animal species. Examples include Creutzfeldt-Jakob disease (CJD) in humans; bovine spongiform encephalopathy (BSE) in cattle; chronic wasting disease (CWD) in deer and elk; and scrapie in sheep and experimental mice (CitationChesebro 2003). There is no test yet available for preclinical diagnosis of transmitted prion disease, nor is there an effective treatment available. In experimental models, certain drug treatments can prolong survival, but early intervention is essential to achieve this effect. We are faced with an urgent need to develop methods both to diagnose and to treat these diseases prior to the onset of clinical symptoms.
TSEs are characterized by vacuolation of the neuropil, neuron loss, activation of astrocytes and microglia, and in some situations the deposition of amyloid fibrils (CitationWells 1993; CitationBudka et al 1995). These pathological changes are thought to depend on a misfolded partially protease-resistant isoform of endogenous prion protein, known as PrPsc or PrP-res (CitationPrusiner 1982; CitationMcKinley et al 1983). The molecular mechanisms underlying the pathology are far from clear (CitationAguzzi and Polymenidou 2004; CitationWeissmann, 2004).
Oral infection is the major route of transmission for many TSEs, including BSE, CWD (CitationBelay et al 2004), transmissible mink encephalopathy (CitationBartz et al 2003), scrapie (CitationAndreoletti et al 2000; Citationvan Keulen et al 2000), and variant CJD (CitationBruce et al 1997). After oral transmission, infectivity passes through the gut to secondary immune tissues and then to the brain (CitationAguzzi and Heikenwalder 2005). Similar to oral infection, experimental intracerebral scrapie inoculation also leads to the secondary lymphoid organs rapidly accumulating infectivity and misfolded prion proteins (CitationFraser and Dickinson 1970; CitationHunter et al 1972).
We hypothesize that genes involved in protein folding and genes that function in the endosome/lysosome system are important cofactors in the misfolding and accumulation of misfolded prion proteins and the propagation of scrapie infectivity. The rationale for this hypothesis include that: 1) prion diseases involve prion protein misfolding (CitationPrusiner 1982; CitationAguzzi 2006), 2) the misfolding of prion proteins is thought to occur on the cell surface or in the endosome/ lysosome pathway (CitationCaughey and Raymond 1991; CitationCaughey et al 1991; CitationCaughey 1993), 3) misfolded prion proteins accumulate in the endosome/lysosome system (McKinley et al 1991), 4) lysosomotropic agents and cysteine protease inhibitors inhibit the scrapie-associated accumulation of prion proteins (CitationDoh-Ura et al 2000), and 5) we have identified a number of genes that function in protein folding and genes that function in the endosome/lysosome system that show increased expression during prion disease (CitationSkinner et al 2006).
In a previous study using cDNA microarrays, we identified 22 genes that showed increased expression in the brains of scrapie-infected mice prior to the onset of clinical symptoms (CitationSkinner et al 2006). One goal of the present study was to expand our gene expression studies using Affymetrix 430A microarrays (Santa Clara, CA), in order to identify additional alterations in expression that occur prior to the onset of clinical symptoms. The Affymetrix 430A microarrays contain over 14,000 unique genes, more than 7000 of which were not represented in the cDNA microarrays in our previous study (CitationSkinner et al 2006). Thus, the Affymetrix microarrays allowed us to identify additional undescribed scrapie-associated gene expression alterations.
Several published TSE gene expression studies have sought to identify alterations in gene expression that occur in the brain. However, given the importance of the lymphoid organs in prion disease transmission and pathogenesis, there is a need to expand gene expression studies to the lymphoid organs. Another goal of the study presented here was to determine in scrapie-infected mice whether genes that show expression alterations in the brain also show expression alterations in spleen tissues.
In this study we used Affymetrix microarrays and identified 67 genes that show alterations in expression in the brains of scrapie-infected mice prior to the onset of clinical symptoms. We confirmed a number of these gene expression alterations using quantitative reverse transcription polymerase chain reaction (qRT-PCR). With qRT-PCR we also determined the time-course of several of these changes. We also determined the time course of astrocytosis and PrPsc accumulation in mice used in these studies, and evaluated protein accumulation of two genes identified GFAP and Gh within the pituitary gland. In addition, we identified 15 genes that showed alterations in gene expression in both spleen and brain tissues from scrapie-infected mice. We further evaluated protein accumulation and localization of one of these genes, anp32a, within the spleen. Our results extend our understanding of prion disease pathogenesis and indicate certain gene products that may serve as surrogate markers for early disease diagnosis.
Methods
Scrapie-infected mice
Three strains of mouse scrapie were examined: ME7, 22L, and RML-Chandler (CitationChandler 1963; CitationZlotnik and Rennie 1963; CitationDickinson 1976). For Affymetrix microarray analyses, we used RNA isolated by the Chesebro lab and shipped to the Skinner lab, from five male C57BL/10 mice that were infected with 50 μl of a 1% scrapie brain homogenate derived from mice at the end stage of scrapie infection. The titer of each inoculum was at least 2 × 108 infectious units/gram brain and exceeded the amount needed to induce disease in 100% of the animals (CitationVorberg et al 2004). RNA was from the same mice used in our previous study (CitationSkinner et al 2006). This included RNA from two mock-infected mice and one ME7- infected, one 22L-infected, and one RML-Chandler-infected mouse, sacrificed at 104 days post-inoculation (dpi).
For the real-time PCR studies a separate set of female C57BL/6 mice was used. For each strain, six C57BL/6 female mice were inoculated intracerebrally with 50 μl of a 1% scrapie brain homogenate derived from mice at the end stage of scrapie infection. The titer of each inoculum was at least 2 × 108 infectious units/gram brain and exceeded the amount needed to induce disease in 100% of the animals (CitationVorberg et al 2004). Six control mice were mock-inoculated with 1% homogenate of brain tissues from uninfected mice. Mock-infected mice served as a control for nonspecific effects induced by the intracerebral injection of brain inoculum. At 46 and 104 dpi (before the onset of clinical symptoms), and at the onset of clinical symptoms at 144 dpi, two mice from each group were sacrificed. Mock- and scrapie-infected mice were sacrificed at the same time of day. Clinical symptoms included kyphosis, dull eyes, flattened stature, weight loss, and ataxia. Total RNA was extracted from whole brain and spleen homogenates using Trizol following the manufacturer’s instructions (Invitrogen), resuspended in 100 μl of RNase free molecular biology grade water (Eppendorf), analyzed by spectrometry and gel electrophoresis for quantity and quality, diluted to 1 μg/μl, and stored at −80 °C.
For the western blot analysis nine female C57BL/6 mice were infected with RML-Chandler strain of scrapie and nine mice were mock-infected as described above and three from each group sacrificed at 45, 100, and 140 dpi.
For the immunohistochemistry staining of GFAP and Gh, one ME7, 22L, RML-Chandler, and one mock-infected C57BL/6 female mouse was infected and sacrificed at 45, 100, and 140 dpi. For anp32a immunohistochemistry three mice were infected with RML-Chandler, and three mock-infected as described above and sacrificed at 45 dpi.
Affymetrix microarrays
Total whole brain RNA from scrapie- and mock-infected mice sacrificed at 104 dpi was reverse transcribed and labeled following protocols provided by Affymetrix. Five Affymetrix Murine 430A chips were hybridized with RNA from two different mock-infected mice and RNA from one ME7-, one 22L-, and one RML-Chandler-infected mouse, following the manufacturer’s instructions. Scan intensity data was extracted and normalized from the Affymetrix output files using the RMAExpress program (http://www.stat.berkeley.edu/~bolstad/RMAExpress/RMAExpress.html). MatchMiner (http://discover.nci.nih.gov/matchminer/index.jsp) was used to convert Affymetrix IDs to corresponding GenBank accession numbers.
Statistical analyses and data mining
We used significance of analysis of microarrays (SAM) with false discovery rates (FDR) and q-values to identify genes that showed differences in expression in scrapie-infected brains compared to mock-infected brains (CitationTusher et al 2001; CitationStorey 2002). We performed a two-class test between results obtained from mock-infected mice and scrapie-infected mice, with data input parameters kept at the default values. We considered for further study those resultant genes that showed a 1.4-fold or greater change in scrapie-infected compared to mock-infected mice and had a q-value of <10%. For simplicity of presentation here, in instances where a gene was represented by more that one Affymetrix probe set, we removed duplicates showing synonymous results.
To determine the function of the proteins encoded by genes showing scrapie-associated alterations in expression, we used published literature, the National Center for Biotechnology Information (NCBI), and the Source (CitationDiehn et al 2003).
Quantitative real-time RT-PCR
For real-time qRT-PCR, we used total RNA from whole brains and spleens. We compared RNA from two mock-infected mice to RNA from six scrapie-infected mice at 46, 104, and 144 dpi. The six scrapie-infected mice included two ME7-, two 22L-, and two RML/Chandler-infected mice. RNA was treated with deoxyribonuclease I (Fermentas, Glen Burnie, MD) to remove genomic DNA contamination and reverse transcribed to cDNA using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA). An M × 3000P qPCR machine (Stratagene, La Jolla, CA) and Platinum SYBR Green qPCR SuperMix-UDG kits (Invitrogen) were used following manufacturers’ instructions. PCR primers were designed using MacVector software (Accelrys Inc., San Diego, CA), and spanned introns. Samples that were not treated with reverse transcriptase were included as a negative control. We previously made a stock of normal mouse brain RNA, treated it with DNAse, reverse transcribed it to cDNA, and purified and used it in each qPCR reaction as a standard to normalize results from experimental samples. The relative cDNA levels were calculated using the 2−ΔΔCT method (CitationLivak and Schmittgen 2001). Actin was used as the reference gene using previously described primers (CitationLengacher et al 2004). PCR products were run on an agarose gel to verify the size of product. For each gene studied, a t-test was performed to verify the significance of the results. The primers and annealing temperatures used, as well as product sizes, are indicated in . Fold change values of less than 1 were converted using the equation −1/fold change, for ease of interpretation.
Table 2 Primers and annealing temperatures used for qRT-PCR studies
Western blot analysis
To detect levels of PrPsc protein accumulation in the spleen and brain, 20% solutions of spleen and brain tissue were prepared in 0.01M TrisHCL pH 7.4, 0.005 M MgCl, treated with DNAse, and sonicated. Tissue lysates were mixed with an equal volume (1 ml) of 20% sarcosyl in 0.01 M Tris HCl pH 7.4 and spun in an ultracentrifuge for thirty minutes in a Beckman ultracentrifuge rotor Type 70.1 (Beckman Coulter, Fullerton, CA) at 5000 RPM. Supernatant were transferred to a new tube and centrifuged at 55000 RPM for two hours. The resultant pellet was resuspended in 1 ml water, sonicated, and then treated with 100 μg/ml proteinase K for 1 hour at 37 °C. Reactions were terminated with 3 mM PMSF. Samples were again be spun at 55000 rpm for one hour and pellets resuspended in 2 × western blot sample buffer, sonicated, and boiled prior to loading on gels. For detection of Prpc and anp32a, brain and spleen tissues were lysed in RIPA buffer (50 mM Tris HCl pH 8, 150 mM NaCl, 1% NP-40, and 0.5% sodium deoxycholate, 0.1% SDS) with protease inhibitors (Sigma P8340; Sigma, St. Louis, MO) and centrifuged at 12000 rpm for 20 minutes. BCA protein assay kit (Pierce Biotechnology, Rockford, IL) was used to determine protein concentrations of resultant supernatant. 6.25 mg/well of protein was loaded for the PrP blot and 12.5 mg/well was used for the anp32a blot. Samples were run using pre-cast polyacrylamide gels in Tris-glycine (4%–12% IM-6000 Invitrogen) and transferred to PVDF membranes (Millipore, Billerica, MA). Blots were probed with antibodies against PrP (SAF-83 from Cayman Labs, or anp32a from AbCam) and then were stripped and re-probed with antibodies directed against Gapdh (Chemicon) and densitometry determined levels of protein accumulation between samples from scrapie-infected and mock-infected mice. Levels of Gapdh were used to normalize each sample.
Immunohistochemistry
Mice were perfused and fixed overnight at 4 °C with freshly made paraformaldehyde. For Gh and GFAP staining tissues were dehydrated in alcohol and paraffin embedded. Sections were treated with antigen retrieval reagent (Diva Decloaker; Biocare, Concord, CA) and then blocked in blocking sniper solution (Biocare) for one hour, and then stained with either GFAP antibodies and counter stained with hematoxylin and eosin, or anti-human Gh antibodies (Biomeda, Foster City, CA) (1:1000) and counter stained with hematoxylin using the Vectastain ABC Kit (Vector Labs, Burlingame, CA). For anp32a staining, after fixation tissues were transferred to phosphate buffer solution (PBS) and stored at 4 °C. Tissues were cut into 50 μm thick sections using a vibratome and blocked with 2% normal goat serum in PBS containing 0.3% Triton- X100, and then stained free floating in 1 ml solution of rabbit anti-anp32a (AbCam) diluted 1:400 and rat anti-mouse-B220 antibodies (BD Pharmingen, San Diego, CA) diluted 1:200 in block solution on a rocking platform over night at 4 °C. Sections were then washed three times for at least 20 minutes with PBS and then incubated with goat anti-rabbit Alexa 488 (Invitrogen) diluted 1:1000 and goat anti-rat Cy3 diluted 1:5000 (Jackson ImmunoResearch, West Grove, PA) and incubated overnight at 4 °C on a rocking platform. Finally sections were again washed then mounted with glycerol-gelatin (Sigma) containing 4 mg/ml n-proply gallate as a fluorophore preservative, and analyzed using a confocal microscope.
Results
Affymetrix microarray analysis of brain gene expression in scrapie-infected mice
We used Affymetrix microarrays and identified 67 genes that showed increased expression in the brains of scrapie-infected mice prior to the onset of clinical symptoms at 104 dpi (). In this study, gene expression was determined in two mock and three scrapie-infected male C57BL/10 mice. The set of scrapie-infected mice included one mouse infected with scrapie strain ME7, one with scrapie strain RML-Chandler, and one with scrapie strain 22L in order to allow us to find scrapie-associated gene expression alterations that occur regardless of strain. A number of the genes we identified have previously been described in related studies of prion disease, primarily at the end stages of disease, and these are cited in (CitationDormont et al 1981; CitationManuelidis et al 1987; CitationDuguid et al 1988; CitationDuguid and Dinauer 1990; CitationDiedrich et al 1993; CitationDuguid and Trzepacz 1993; CitationWilliams et al 1994; CitationDandoy-Dron et al 1998; CitationBaker et al 1999, Citation2002, Citation2004; CitationDoh-ura et al 2000; CitationGayrard et al 2000; CitationKopacek et al 2000; CitationRiemer et al 2000, Citation2004; CitationBaker and Manuelidis 2003; CitationRangon et al 2003; CitationZhang et al 2003; CitationBooth et al 2004a, Citation2004b; CitationBrown et al 2004; CitationLu et al 2004; CitationSchultz et al 2004; CitationXiang et al 2004; CitationViguie et al 2004; CitationGreenwood et al 2005; CitationRodriguez et al 2006; CitationSkinner et al 2006). Eighteen of the genes we identified have not been previously reported in prion disease studies (). The genes identified function in multiple cellular processes including: immunity, the endosome lysosome system, the cytoskeleton, and hormone activity. Many of the genes function in multiple cellular processes, but are listed in a single functional category for ease of presentation.
Table 1 Scrapie-associated expression alterations in brain at 104 dpi using Affymetrix microarrays
Onset of pathology
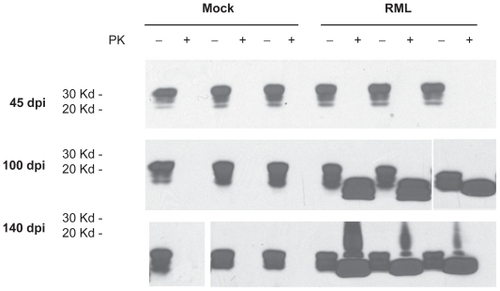
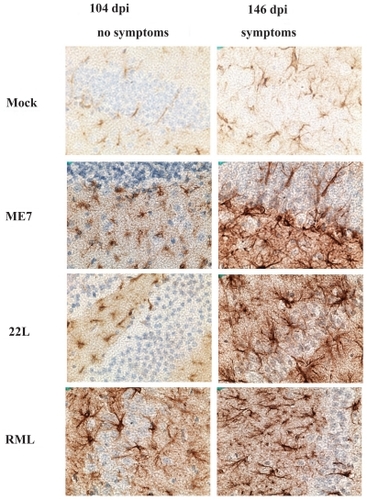
In order to establish when pathological PrPsc accumulation developed in the scrapie-infected mice, we performed western blot analysis of brain and spleen extracts from three mice infected with RML-Chandler strain of scrapie and three mock-infected mice at 45, 100, and 140 dpi. In the brain, PrPc was readily detectible at all time points examined. No proteinase K resistant PrPsc was detectible at 45 dpi, but was readily detectible at 100 and 140 dpi (). This correlated with detection of increased GFAP immunoreactivity in astrocytes throughout the brain in mice infected with Me7, 22L, and RML-Chandler strains of scrapie at 104 and 146 dpi (). In addition, our western blot analysis of PrPc and PrPsc in spleen tissues from the same scrapie-infected mice revealed that the proteinase K digestion was incomplete, but nonetheless showed abundant PrPc and PrPsc at 45, 100, and 140 dpi (data not shown).
Figure 1 Prion protein and proteinase K resistant prion protein accumulation in brains from mock and RML-Chandler scrapie-infected mice at 45, 100, and 140 days post-infection. Western blot analysis showing prion protein antibody staining (SAF83) of protein extracts that were untreated or treated with proteinase K.
Appendix 2 GFAP immunoreactivity in scrapie-infected and mock-infected mice at 104 and 146 dpi. Hippocampus region of the brain from sections from ME7, 22L, and RML-Chandler-infected mice stained with GFAP antibodies (brown) and counterstained with hematoxylin (blue). Note the increase in GFAP immunoreactivity in the scrapie compared to mock-infected mice.
qRT-PCR confirmation of microarray results
We used quantitative RT-PCR (qRT-PCR) to confirm a subset of the scrapie-associated alterations in expression of the genes identified in our Affymetrix study that have not previously been described in association with TSEs, and to determine the time course over which these alterations occur. Twelve genes were selected that were representative of each functional category of genes identified in the Affymetrix microarray study: immunity, lysosome, ubiquitin/proteosome, and other. The genes evaluated were Usp18, Olfml3, Ifit1, Ifit3, Ifitm3, Ifit27, Rtp4, Stat1, Trem2, Grn, H2-T2, and Anp32a. Hormone genes were also evaluated and discussed in the next section. We evaluated gene expression in at three time points post-infection: prior to the onset of clinical symptoms (at 44 and 104 dpi) and at the time when symptoms became detectable (at 144 dpi). These time points were selected because they are similar to as those used in our previous study (CitationSkinner et al 2006), and thus allow the two studies to be correlated. Gene expression levels were determined at each time point in two mock-infected and six scrapie-infected female C57BL/6 mice. The set of scrapie-infected mice included two infected with strain ME7, two with strain 22L, and two with strain RML-Chandler. As controls in this study, we used two genes: Atp1b1, because it showed decreased expression and GFAP, because it showed increased expression in the brains of scrapie-infected mice in our previous studies (CitationSkinner et al 2006). Primers, annealing temperatures, and product sizes are presented in . We show the average expression from all three strains of scrapie-infected mice relative to mock-infected mice in , and we present in our more detailed results for each scrapie strain: ME7, 22L, and RML-Chandler. In this study, none of the genes showed significantly altered expression at 46 dpi. Five of the 12 genes showed significantly increased expression at 104 dpi. These genes were Usp18, Olfml3, Ifit1, Ifit3, and Ifi27. At the onset of clinical symptoms (144 dpi), 11 of the 12 genes (all but Anp23a) showed significant increased expression in the brain of scrapie-infected mice relative to mock-infected mice. Overall, although not statistically significant in every case, all 12 experimental genes showed higher expression in scrapie-infected mice compared to mock-infected mice at some point in time. Although none of the experimental genes showed significant alteration in the brain at the very early time point 46 dpi, interestingly, the control gene Atp1b1 showed significantly decreased expression at 46 dpi, demonstrating that alterations in Atp1b1 occur very early in the course of scrapie infection.
Table 3 Confirmation of scrapie-associated gene expression alterations using qRT-PCR
Appendix 1 Brain gene expression in mice infected with ME7, 22L, and RML-Chandler strains of scrapie vs. mock-infected mice using qRT-PCR
Alterations in hormone expression in scrapie-infected mice
Four of the genes that showed increased expression in the brain of scrapie-infected mice prior to the onset of clinical symptoms encode hormones expressed primarily in the pituitary gland. These genes were Cga, Gh, Prl, and Pomc1. To confirm these increases and to determine the time course over which these increases in hormone expression take place, we performed qRT-PCR and evaluated gene expression in scrapie-infected and mock-infected mice at 46, 104, and 144 dpi. Results shown in depict the average of all three scrapie strains. Results from individual strains are presented in . Gh showed significant increased expression at the very early time point 46 dpi. Gh, Prl, and Pomc1 showed increased expression at 104 and 144 dpi. In this study, Cga did not show significant increased expression at any time point using qRT-PCR.
Table 4 Hormone and hormone regulator gene expression in scrapie-infected vs mock-infected mice using qRT-PCR
We then used qRT-PCR to determine whether the upstream regulators of these hormones might themselves be upregulated in scrapie-infected mice. We evaluated the expression of Crh, Ghrh, Trh, and Gnrh1, which induce the expression and secretion of hormones in the pituitary gland. None of these genes showed significant expression alterations in presymptomatic mice. However, Crh showed increased expression at the onset of clinical symptoms (144 dpi) (). We also determined whether other hormone genes that are primarily expressed in the pituitary gland were altered in pre-symptomatic scrapie-infected mice by evaluating expression levels of Lhb, Tshb, and Fshb in scrapie-and mock-infected mice. These gene products bind to Cga (glycoprotein hormones alpha subunit) to form luteinizing hormone, thyroid-stimulating hormone, and follicle-stimulating hormone. None of these genes were significantly altered in scrapie-infected mice (). These results confirm the findings from our Affymetrix microarray study, which showed increased expression of Gh, Prl, and Pomc1 in the brains of scrapie-infected mice. The results also show that not all hormone-encoding genes that are primarily expressed in the pituitary gland are altered in scrapie-infected mice. The increased expression of hormone genes identified in our study is not likely due to increased expression of upstream regulators of hormones but rather by some other mechanism.
Increases GFAP and Gh in anterior pituitary gland of scrapie-infected mice
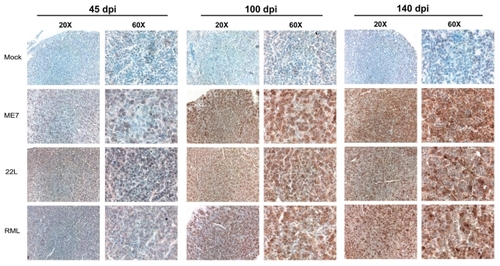
Two of the genes that were found to be upregulated in scrapie-infected mice were GFAP and Gh. We stained pituitary glands from ME7, 22L, and RML Chandler-infected mice and mock-infected mice at 45, 100, and 145 dpi with GFAP and Gh antibodies. Interestingly, both GFAP and Gh antibodies showed increased immunoreactivity in glandular hormone secreting cells of the anterior pituitary gland ( and ).
Figure 2 GFAP antibody staining in the anterior pituitary gland (adenohypophysis) from scrapie- and mock-infected mice at 45, 100, and 140 days post-infection. Sections were stained with GFAP antibodies (brown) and counterstained with hematoxylin (blue) and eosin (pink). Images were collected with 20 × and 60 × objectives as indicated. Note increased GFAP immunoreactivity in glandular cells of anterior pituitary in sections from scrapie-infected mice relative to mock-infected mice.
Figure 3 Growth hormone (Gh) antibody staining in the anterior pituitary gland (adenohypophysis) from scrapie- and mock-infected mice at 45, 100, and 140 days post-infection. Images show sections stained with Gh antibodies (brown) and counterstained with haematoxylin (blue). Images were collected with 20 × and 60 × objectives as indicated. Note increased Gh immunoreactivity in glandular cells of anterior pituitary in sections from scrapie-infected mice relative to mock-infected mice.
Alterations in gene expression in the spleen of scrapie-infected mice
Experimental intracerebral scrapie inoculation leads to the rapid accumulation of infectivity and PrPsc in secondary lymphoid organs including the spleen (CitationFraser and Dickinson 1970; CitationHunter et al 1972). To analyze gene expression in the spleen of scrapie-infected mice, we used qRT-PCR to evaluate relative expression levels of 22 genes that in our current or previous microarray studies (CitationSkinner et al 2006) showed scrapie-associated expression alterations in the brain. These genes were selected 1) because they encode proteins that function either in protein folding or in the endosome/lysosome system (where misfolded prion proteins accumulate), 2) because they function in immunity (a major functional group of genes that show altered expression in the brain during prion disease), or 3) because they were novel genes that have never been described in association with prion diseases. These genes were Hspa8, Hspa12a, Hspa4, Ctsz, Ctss, HexB, H2-T23, Ifit1, Ifit3, Ifitm3, Ifi27, Rtp4, Cd68, Gh, Grn, Stat1, Usp18, Trem2, Olfml3, and Anp32a. We also included GFAP and Atp1b1 because in our previous studies they showed increased and decreased expression, respectively, in the brain early in the course of scrapie infection (CitationSkinner et al 2006). We evaluated gene expression in the spleen at the same time points as in our brain studies: at two time points prior to the onset of clinical symptoms (at 46 and 104 dpi) and at the time when clinical symptoms appeared in the mice (at 144 dpi).
The results showed that all 22 genes evaluated were expressed in the spleen. At 46 dpi, two of the 22 genes – Anp32a and Grn – showed significant increases in expression in the spleens of scrapie-infected mice compared to mock-infected mice (). These two genes also showed significantly increased expression at the later time points, 104 and 144 dpi. At 104 dpi, in addition to Anp32a and Grn, 12 other genes showed significantly increased expression relative to mock-infected mice. These genes were Ctsz, HexB, Ctss, Hspa4, Hspa8, Hspa12a, Ifit1, Ifit3, Ifitm3, Cd68, Ifi27, and Olfml3 (). At the onset of clinical symptoms, at 144 dpi, these 14 genes continued to show significant increased levels of expression, and were joined by H2-T23 (). The remaining genes, Gh, Stat1, Usp18, Rtp4, Gfap, Trem2, and Atp1b1, were expressed in the spleen, but none showed significantly altered expression at any time point (). shows the results from averaging the expression of all three scrapie strains relative to mock-infected mice. shows results obtained for each strain individually. These results demonstrate that prior to the onset of clinical symptoms, scrapie induces multiple alterations in gene expression that occur in both the spleen and brain.
Table 5 Spleen gene expression in scrapie-infected vs mock-infected mice at 46, 104, and 144 dpi
Appendix 3 Spleen gene expression in mice infected with ME7, 22L, and RML-Chandler strains of scrapie vs. mock-infected mice using qRT-PCR
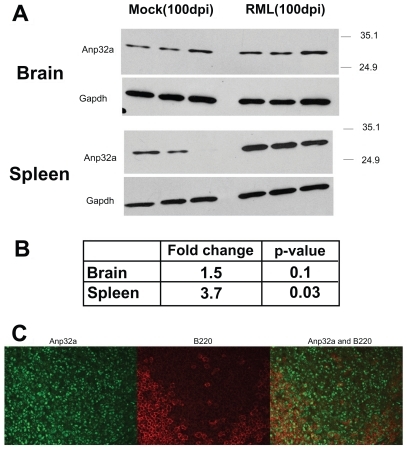
In order to determine if anp32a which showed very early increased expression in spleens of scrapie-infected mice also showed increased protein accumulation in the spleen or brain of scrapie-infected mice, we performed western blot analysis using brains and spleens from three mice infected with RML/ Chandler strain of scrapie and three mock-infected mice sacrificed at 100 dpi. As shown in ure 4, we found anp32a protein accumulated 3.7 times more in spleens from scrapie-infected mice compared to mock-infected mice (p = 0.03). In the brain, anp32a showed 1.5 times more anp32a protein in scrapie-infected compared to mock-infected mice, but this trend was not significant (p = 0.1).
In order to localize anp32a protein expression within the spleen of scrapie-infected mice, we stained spleen sections from RML/Chandler scrapie-infected mice with antibodies directed against anp32a and the B cell marker B220. The results showed that anp32a was expressed in B cells as well as other cell types ()
Figure 4 Anp32a protein accumulation in brain and spleen of scrapie-and mock-infected mice. A) Western blot results from three mock and three scrapie-infected mice at 100 days post-infection stained with anp32a antibodies, and stripped and reprobed with Gapdh antibodies. B) Densitometry analysis of western blot results showing relative amounts of anp32a accumulation in scrapie verses mock-infected mice. C) Anp32a (green) and B220 (red) antibody staining in spleen tissue from scrapie-infected mouse. Note: B220 is a B cell marker.
Discussion
These studies shed light on the complex molecular events that occur during prion disease and identify genes whose further study may yield new insights into TSE pathogenesis and transmission. We identified 67 genes that show altered expression in the brain and 15 genes that show altered expression in both the spleen and brain of scrapie-infected mice. All of the scrapie-associated alterations in gene expression in the brain, and 14 out of 15 in the spleen, occurred prior to the development of clinical symptoms and therefore may lead to the development of ante-mortem tests for TSEs.
Fourteen genes showed increased expression in the spleens of scrapie-infected mice prior to the onset of clinical symptoms. These same genes showed increased expression in the brains of scrapie infected mice as presented here or in our previous studies (CitationSkinner et al 2006). Because these increases in gene expression occur in both the brain and the spleen prior to the onset of disease, and because prion infectivity and PrPsc accumulates in both the spleen and brain of scrapie infected mice, these genes may be important contributors to the mechanism by which prion proteins misfold and accumulate and to the dissemination of TSE infectivity.
Importantly, all seven of the genes evaluated in the spleen that function in protein folding or in the endosome/lysosome system showed increased expression in scrapie-infected mice relative to mock-infected mice. These genes were Hspa8, Hspa4, Hspa12a, Cd68, Ctsz, Ctss, and HexB. These results support our hypothesis that genes that function in protein folding or in the endosome/lysosome system contribute to prion disease pathogenesis and are important cofactors in the misfolding and accumulation if prion proteins. Most of the genes involved in immunity also showed alterations in expression in the spleens of scrapie-infected mice. These genes were Grn, Ifit1, Ifit3, fitm3, Ifi27, and H2-T23. In addition, Anp32a, which functions in signal transduction, and Olfml3, a gene of unknown function, were also found to show increased expression in spleens of scrapie-infected compared to mock-infected mice. Gene alterations in the spleen may prove useful as markers for the preclinical diagnosis of prion diseases. It will be particularly important to identify which cell types alterations occur in and whether these alterations also occur in cells in the blood.
Some genes differ with regard to the time post-infection at which altered expression was first observed in the spleen compared to brain. Grn and Anp32a showed increased expression in the spleen at 46 dpi, but did not show altered expression in the brain until 104 dpi. Gh showed increased expression, and Atb1b1 showed decreased expression in the brains of scrapie-infected mice at 46, 104, and 144 dpi, but neither showed significant alterations in expression at any of these time points in the spleen. Olfml3 and Ifit1 showed altered expression at 104 dpi in both brain and spleen tissues. These similarities and differences in the time course of alterations between the spleen and brain tissues might be attributed to similar and different pathological changes that are occurring in each of these tissue compartments during the course of disease.
It is unclear why there was a delay in detection upregulated genes in our qRT-PCR study relative to our microarray study, but may be due to differences in the sex and stains of mice used in the studies. A set of male C57BL/10 mice was used for the microarray studies, and a different set of female C57BL/6 mice was used for the qRT-PCR studies. Differences may also be due to small sample numbers. Importantly, because our qRT-PCR results did show that 11 of 12 genes examined showed significant increased expression at 146 dpi, these results confirm that these 11 genes show scrapie-associated expression alterations and validate the microarray results.
Very little is known about the molecular changes that take place very early after TSE infection. Four genes in this study showed altered expression at the very early time point, 46 dpi. In the brain, we observed significantly decreased expression of ATPase Na+/K+ transporting beta 1 polypeptide (Atp1b1) and increased expression in Gh. In the spleen acidic leucine-rich nuclear phosphoprotein 32 family member A (Anp32a) and granulin (Grn) showed significant increased expression. Atb1b1 is an intrinsic membrane protein that functions in the active transport of Na+ and K+ ions in most animal cells, is important for transmission of action potentials in neurons, and also is associated with caveolae and intracellular signal transduction events (CitationKaplan 2002; CitationLiu and Askari 2006). Gh, in addition to stimulating growth, also functions in an autocrine and paracrine fashion in reproduction processes, immunity, tooth development, retinal development, and placental function (CitationHarvey et al 2003; CitationHarvey and Hull 2003). Anp32a is thought to function in signal transduction and interacts with the cytoskeleton (CitationOpal et al 2003; CitationMatilla and Radrizzani 2005), and has been implicated in other neurodegenerative diseases. For example, its expression is decreased in spinocerebellar ataxia type 3 (CitationEvert et al 2003), and it binds to and colocalizes with mutant ataxin-1, the protein that causes spinocerebellar ataxia type 1 (CitationMatilla et al 1997). Full length Grn gets processed into a variety of 6 kDa peptides and full length protein and peptides are important for, regulation of cell growth, normal development, wound healing, and tumorigenesis. Interestingly, mutations in Grn have recently been discovered to cause frontotemporal dementia linked to chromosome 17 (FTD-17) (CitationBaker et al 2006; CitationCruts et al 2006; CitationMukherjee et al 2006). Additional studies are needed to expand our understanding of the contributions to TSE disease pathogenesis and progression of Gh, Atp1b1, Anp32a and Grn, as well as the other scrapie-induced alterations in gene expression identified in this study that occur later in the progression of the disease. In addition, we evaluated only 22 genes at the very early time-point 46 days post-infection. Additional large scale microarray studies of tissues from scrapie-infected compared to mock-infected mice at very early time-points post infection are warranted.
Our studies also found that the pituitary hormones prolactin (Prl), Gh, and pro-opiomelanocortin-alpha (Pomc1) showed increased expression in the brains of scrapie-infected mice prior to the development of clinical symptoms. Pomc1 encodes a 235 amino acid protein that gets processed to produce at least eight peptides including: adrenocorticotropic hormone (ACTH), lipotropins, endorphins, and melanocyte-stimulating hormones (MSHs). The scrapie-associated upregulation of these genes was not associated with a concurrent increase in the expression in upstream regulators of these hormones, Crh, Gnrh1, Trh, and Ghrh, indicating that some other mechanism is responsible for the upregulation pituitary hormones identified in our study. Previous studies in hamsters showed that scrapie strain 139H, but not strain 263K, highly impacts the pituitary gland, inducing neuropathology including vacuolization, atrophy, cytoplasmic vesicles, and hypertrophy (CitationYe and Carp 1996). This neuropathology in the pituitary gland is associated with hyperplasia and hypertrophy of islets of Langerhan cells, thyroid glands, adrenal glands, liver, and kidneys (CitationYe and Carp 1996). In addition, increased plasma levels of Gh, ACTH, and cortisol are evident during natural sheep scrapie (CitationGayrard et al 2000; CitationViguie et al 2004). Taken together, these studies indicate that alterations in the expression of pituitary hormones may be responsible for some of the clinical symptoms observed during scrapie infection in mice, hamsters, and sheep.
This study identified alterations in gene expression that occur in three strains of scrapie in mice. Mice infected intracerebrally with the scrapie strains ME7, 22L and RML-Chandler differ in incubation period, in neuropathology, and in disease associated alterations in gene expression (CitationDickinson 1976; CitationBruce and Fraser 1991; CitationKascsak et al 1991; CitationBooth et al 2004a; CitationSkinner et al 2006). Alterations that occur regardless of strain and in multiple TSEs are more likely to play a critical role in TSE pathogenesis and disease progression, whereas alterations in gene expression that do not occur in all TSEs or in only some strains are more likely to be associated with strain-specific neuropathological changes.
These studies show that as few as five mice, two mock-infected and three scrapie-infected (with scrapie strain ME7, 22L, and RML-Chandler, respectively), are sufficient to identify alterations in gene expression that occur during the course of scrapie infection in C57BL mice. Our analysis identified 67 gene expression alterations in scrapie-infected mice relative to mock-infected mice. These alterations were verified by the fact that 1) most of the genes identified in our study have been described by us or others in related prion disease studies, 2) the novel gene alterations described here were confirmed in the brain a separate set of scrapie-infected mice using qRT-PCR, and 3) several of these genes were also found to show significant increased expression in spleen tissues from scrapie-infected mice relative to mock-infected mice. Thus, Affymetrix microarray studies using only five mice can prove useful for the identification of gene expression alterations in scrapie-infected mice. At the same time, the small sample size with the microarray study likely limited the total amount of gene alterations identified. Additional studies with increased numbers of mice may yield more genes that show significant scrapie-associated changes in gene expression.
Although many of the genes identified in this study showed relatively large (greater than two-fold) changes in expression relative to mock-infected mice, many other genes showed relatively low (less than two-fold) changes. Relatively low-fold changes in gene expression may lead to substantial effects in the brain that are important to prion disease pathogenesis. Also, because our studies used RNA from total brain tissue including multiple cell types from multiple regions of the brain, we anticipate that gene alterations that occur in a single cell type or only in a particular region of the brain will show a low fold change in this analysis due to dilution by mRNA from surrounding cells or brain regions in which the particular gene alteration is not occurring. Thus, for gene expression analyses involving complex tissues such as brain, low fold changes in gene expression may indicate large fold changes in a subset of cells or cells in a particular region of the tissue and should not be overlooked.
Future studies are needed to determine whether the scrapie-associated alterations identified in mice also occur in other animals and humans with TSEs, and whether the alterations are specific to prion disease. We also need to determine what the range is of normal variation in expression levels among individuals. To our knowledge, the decrease in erythroid differentiation-related factor (EDRF) expression is the only other nonneuronal alteration in gene expression that is associated with prion diseases described to date (CitationMiele et al 2001). Decreased EDRF expression levels were found in spleen, bone marrow, and blood of scrapie-infected rodents, and were also found in tissues from BSE-infected cows and scrapie-infected sheep. As researchers attempted to translate these findings to a diagnostic assay for human TSEs, they discovered that EDRF expression levels in blood are highly variable amongst individual humans, making it difficult to use EDRF expression levels as a marker for disease (CitationGlock et al 2003). We are hopeful that some of the genes identified in this study that show scrapie-associated changes, will show similar disease-associated changes in humans and show low variability in expression between humans and thus be good surrogate markers for the early diagnosis of prion disease in humans and animals.
Conclusions
We identified a set of genes that show altered expression prior to the onset of clinical symptoms in spleen and brain tissues from scrapie-infected mice. These studies take us several steps forward in our understanding of the molecular events underlying scrapie pathogenesis. Some of the genes identified in this study may serve as surrogate markers for prion disease diagnosis and lead to the identification of targets for drug therapies.
Disclosure
HK performed scrapie infections of female mice, sacrificed and dissected mice, processed tissues, performed qRT-PCR analyses and GFAP and Gh immunohistochemistry; GS performed Affymetrix microarray analyses and helped with scrapie infections; TB performed Anp32a immunohistochemistry; BC provided RNA from male scrapie-infected mice, and helped with manuscript preparation; RR did the scrapie infections and extracted the RNA from male mice used for the microarray studies; and PS was the project leader, oversaw experimentation, and drafted this manuscript.
We thank Kimberly Meade-White and Anne Ward for training with scrapie infections; H. Brent Clark for assistance with histopathology analysis; Hayet Abbassi for preparing the RNAs used in Affymetrix hybridizations and reviewing this manuscript; Karin Matchett for editing this manuscript; and the UMN Biomedical Genomics Center for Affymetrix hybridization and data analysis support. This work was supported by grants from the University of Minnesota Biomedical Genomics Center.
References
- AguzziAHeikenwalderM2005Prions, cytokines, and chemokines: a meeting in lymphoid organsImmunity221455415723804
- AguzziAPolymenidouM2004Mammalian prion biology: one century of evolving conceptsCell1163132714744440
- AguzziA2006Prion diseases of humans and farm animals: epidemiology, genetics, and pathogenesisJ Neurochem9717263916805779
- AndreolettiOBerthonPMarcD2000Early accumulation of PrP(Sc) in gut-associated lymphoid and nervous tissues of susceptible sheep from a Romanov flock with natural scrapieJ Gen Virol81Pt 1231152611086143
- BakerCALuZYManuelidisL2004Early induction of interferon-responsive mRNAs in Creutzfeldt-Jakob diseaseJ Neurovirol10294014982726
- BakerCALuZYZaitsevI1999Microglial activation varies in different models of Creutzfeldt-Jakob diseaseJ Virol7350899710233972
- BakerCAManuelidisL2003Unique inflammatory RNA profiles of microglia in Creutzfeldt-Jakob diseaseProc Natl Acad Sci U S A100675912525699
- BakerCAMartinDManuelidisL2002Microglia from Creutzfeldt-Jakob disease-infected brains are infectious and show specific mRNA activation profilesJ Virol76109051312368333
- BakerMMackenzieIRPickering-BrownSM2006Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17Nature442916916862116
- BartzJCKincaidAEBessenRA2003Rapid prion neuroinvasion following tongue infectionJ Virol775839112477862
- BelayEDMaddoxRAWilliamsES2004Chronic wasting disease and potential transmission to humansEmerg Infect Dis109778415207045
- BoothSBowmanCBaumgartnerR2004aMolecular classification of scrapie strains in mice using gene expression profilingBiochem Biophys Res Commun32513394515555574
- BoothSBowmanCBaumgartnerR2004bIdentification of central nervous system genes involved in the host response to the scrapie agent during preclinical and clinical infectionJ Gen Virol85Pt 1134597115483264
- BrownARWebbJRebusS2004Identification of up-regulated genes by array analysis in scrapie-infected mouse brainsNeuropathol Appl Neurobiol305556715488032
- BruceMEFraserH1991Scrapie strain variation and its implicationsCurr Top Microbiol Immunol172125381810707
- BruceMEWillRGIronsideJW1997Transmissions to mice indicate that ‘new variant’ CJD is caused by the BSE agentNature3894985019333239
- BudkaHAguzziABrownP1995Neuropathological diagnostic criteria for Creutzfeldt-Jakob disease (CJD) and other human spongiform encephalopathies (prion diseases)Brain Pathol5459668974629
- CaugheyBRaymondGJErnstD1991N-terminal truncation of the scrapie-associated form of PrP by lysosomal protease(s): implications regarding the site of conversion of PrP to the protease-resistant stateJ Virol6565976031682507
- CaugheyBRaymondGJ1991The scrapie-associated form of PrP is made from a cell surface precursor that is both protease- and phospholipase-sensitiveJ Biol Chem26618217231680859
- CaugheyB1993Scrapie associated PrP accumulation and its prevention: insights from cell cultureBr Med Bull49860728137133
- ChandlerRL1963Experimental scrapie in the mouseRes Vet Sci4276
- ChesebroB2003Introduction to the transmissible spongiform encephalopathies or prion diseasesBr Med Bull6612014522845
- CrutsMGijselinckIvan der ZeeJ2006Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21Nature442920416862115
- Dandoy-DronFGuilloFBenboudjemaL1998Gene expression in scrapie. Cloning of a new scrapie-responsive gene and the identification of increased levels of seven other mRNA transcriptsJ Biol Chem273769179516475
- DickinsonAG1976Scrapie in sheep and goatsFront Biol4420941821790
- DiedrichJFCarpRIHaaseAT1993Increased expression of heat shock protein, transferrin, and beta 2- microglobulin in astrocytes during scrapieMicrob Pathog15168412623
- DiehnMSherlockGBinkleyG2003SOURCE: a unified genomic resource of functional annotations, ontologies, and gene expression dataNucleic Acids Res312192312519986
- Doh-UraKIwakiTCaugheyB2000Lysosomotropic agents and cysteine protease inhibitors inhibit scrapie- associated prion protein accumulationJ Virol744894710775631
- Doh-uraKMekadaEOgomoriK2000Enhanced CD9 expression in the mouse and human brains infected with transmissible spongiform encephalopathiesJ Neuropathol Exp Neurol597748511005258
- DormontDDelpechBDelpechA1981[Hyperproduction of glial fibrillary acidic protein (GFA) during development of experimental scrapie in mice]C R Seances Acad Sci III2935366796203
- DuguidJTrzepaczC1993Major histocompatibility complex genes have an increased brain expression after scrapie infectionProc Natl Acad Sci U S A9011477678332
- DuguidJRDinauerMC1990Library subtraction of in vitro cDNA libraries to identify differentially expressed genes in scrapie infectionNucleic Acids Res182789922339063
- DuguidJRRohwerRGSeedB1988Isolation of cDNAs of scrapie-modulated RNAs by subtractive hybridization of a cDNA libraryProc Natl Acad Sci U S A855738422456582
- EvertBOVogtIRVieira-SaeckerAM2003Gene expression profiling in ataxin-3 expressing cell lines reveals distinct effects of normal and mutant ataxin-3J Neuropathol Exp Neurol6210061814575237
- FraserHDickinsonAG1970Pathogenesis of scrapie in the mouse: the role of the spleenNature22646234985820
- GayrardVPicard-HagenNGrinoM2000Major hypercorticism is an endocrine feature of ewes with naturally occurring scrapieEndocrinology1419889410698174
- GlockBWinterMRennhoferSO2003Transcript level of erythroid differentiation-related factor, a candidate surrogate marker for transmissible spongiform encephalopathy diseases in blood, shows a broad range of variation in healthy individualsTransfusion4317061014641867
- GreenwoodADHorschMStengelA2005Cell line dependent RNA expression profiles of prion-infected mouse neuronal cellsJ Mol Biol34948750015896347
- HarveySHullK2003Neural growth hormone: an updateJ Mol Neurosci2011412663929
- HarveySKakebeekeMMurphyAE2003Growth hormone in the nervous system: autocrine or paracrine roles in retinal function?Can J Physiol Pharmacol813718412769229
- HunterGDKimberlinRHMillsonGC1972Absence of eclipse phase in scrapie miceNat New Biol2353124623142
- KaplanJH2002Biochemistry of Na,K-ATPaseAnnu Rev Biochem715113512045105
- KascsakRJRubensteinRCarpRI1991Evidence for biological and structural diversity among scrapie strainsCurr Top Microbiol Immunol172139521725770
- KopacekJSakaguchiSShigematsuK2000Upregulation of the genes encoding lysosomal hydrolases, a perforin-like protein, and peroxidases in the brains of mice affected with an experimental prion diseaseJ Virol74411710590130
- LengacherSMagistrettiPJPellerinL2004Quantitative rt-PCR analysis of uncoupling protein isoforms in mouse brain cortex: methodological optimization and comparison of expression with brown adipose tissue and skeletal muscleJ Cereb Blood Flow Metab24780815241186
- LiuLAskariA2006Beta-subunit of cardiac Na+-K+-ATPase dictates the concentration of the functional enzyme in caveolaeAm J Physiol Cell Physiol291C5697816624992
- LivakKJSchmittgenTD2001Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) MethodMethods25402811846609
- LuZYBakerCAManuelidisL2004New molecular markers of early and progressive CJD brain infectionJ Cell Biochem936445215660413
- ManuelidisLTesinDMSklaviadisT1987Astrocyte gene expression in Creutzfeldt-Jakob diseaseProc Natl Acad Sci U S A845937412441399
- MatillaAKoshyBTCummingsCJ1997The cerebellar leucine-rich acidic nuclear protein interacts with ataxin-1Nature38997489353121
- MatillaARadrizzaniM2005The Anp32 family of proteins containing leucine-rich repeatsCerebellum471815895553
- McKinleyMPBoltonDCPrusinerSB1983A protease-resistant protein is a structural component of the scrapie prionCell3557626414721
- McKinleyMPTaraboulosAKenagaL1961Ultrastructural localization of scrapie prion proteins in cytoplasmic vesicles of infected cultured cellsLab Invest65622301684401
- MieleGMansonJClintonM2001A novel erythroid-specific marker of transmissible spongiform encephalopathiesNat Med7361411231637
- MukherjeeOPastorPCairnsNJ2006HDDD2 is a familial frontotemporal lobar degeneration with ubiquitin-positive, tau-negative inclusions caused by a missense mutation in the signal peptide of progranulinAnn Neurol603142216983685
- OpalPGarciaJJPropstF2003Mapmodulin/leucine-rich acidic nuclear protein binds the light chain of microtubule-associated protein 1B and modulates neuritogenesisJ Biol Chem27834691912807913
- PrusinerSB1982Novel proteinaceous infectious particles cause scrapieScience1982216136446801762
- RangonCMHaikSFaucheuxBA2003Different chromogranin immunoreactivity between prion and a-beta amyloid plaqueNeuroreport14755812692477
- RiemerCNeidholdSBurwinkelM2004Gene expression profiling of scrapie-infected brain tissueBiochem Biophys Res Commun3235566415369787
- RiemerCQueckISimonD2000Identification of upregulated genes in scrapie-infected brain tissueJ Virol7410245811024157
- RodriguezAPerez-GraciaEEspinosaJC2006Increased expression of water channel aquaporin 1 and aquaporin 4 in Creutzfeldt-Jakob disease and in bovine spongiform encephalopathy-infected bovine-PrP transgenic miceActa Neuropathol (Berl)1125738516871401
- SchultzJSchwarzANeidholdS2004Role of interleukin-1 in prion disease-associated astrocyte activationAm J Pathol165671815277240
- SkinnerPJAbbassiHChesebroB2006Gene expression alterations in brains of mice infected with three strains of scrapieBMC Genomics711416700923
- StoreyJ2002A direct approach to false discovery ratesJ R Stat Soc Ser B Stat Methodol6447998
- TusherVGTibshiraniRChuG2001Significance analysis of microarrays applied to the ionizing radiation responseProc Natl Acad Sci U S A9851162111309499
- van KeulenLJSchreuderBEVromansME2000Pathogenesis of natural scrapie in sheepArch Virol Suppl16577111214935
- ViguieCChilliardYGayrardV2004Alterations of somatotropic function in prion disease in sheepJ Endocrinol1834273515531730
- VorbergIRainesAPriolaSA2004Acute formation of protease-resistant prion protein does not always lead to persistent scrapie infection in vitroJ Biol Chem279292182515133048
- WeissmannC2004The state of the prionNat Rev Microbiol28617115494743
- WellsGA1993Pathology of nonhuman spongiform encephalopathies: variations and their implications for pathogenesisDev Biol Stand806198270117
- WilliamsAELawsonLJPerryVH1994Characterization of the microglial response in murine scrapieNeuropathol Appl Neurobiol2047558208340
- XiangWWindlOWunschG2004Identification of differentially expressed genes in scrapie-infected mouse brains by using global gene expression technologyJ Virol78110516015452225
- YeXCarpRI1996Histopathological changes in the pituitary glands of female hamsters infected with the 139H strain of scrapieJ Comp Pathol1142913048762587
- ZhangYSpiessEGroschupMH2003Up-regulation of cathepsin B and cathepsin L activities in scrapie-infected mouse Neuro2a cellsJ Gen Virol84Pt 822798312867662
- ZlotnikIRennieJC1963Further observations on the experimental transmission of scrapie from sheep and goats to laboratory miceJ Comp Pathol731506214003830