Abstract
Background
The aim of this study was to compare the recovery of three putative periodontal pathogens from periodontal lesions in samples using paper points inserted to different depths of the lesions.
Methods
Twenty 6–8 mm deep periodontal lesions with bleeding on probing were studied. Microbial samples were obtained using paper points inserted to three different depths of the lesions: orifice of lesion; 2 mm into the lesion; and to the base of lesion. Culturing was used for recovery and identification of Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, and Prevotella intermedia.
Results
The recovery of each of the three putative periodontal pathogens was similar following sampling at the various depths of the lesions.
Conclusions
The findings may be explained by the fact that the paper points become saturated as they pass through the orifice of the lesion. Absorption of microorganisms will therefore primarily occur at the orifice. It is also conceivable that the pathogens may be present in similar proportions throughout the various depths of the periodontal lesions.
Introduction
Periodontal disease is a multifactoral infection elicited by a complex microbiota in the gingival crevicular area.Citation1–Citation3 A valid sampling technique is essential to identify and locate the different components of the microbiota of periodontal lesions.
Microbial sampling using endodontic, absorbent paper points inserted to the depth of the periodontal lesion seems to be the most commonly employed sampling method.Citation4,Citation5 A concern with this technique is that the samples may not reflect the microbiota of the deeper part of the lesion. The microbiota in the deeper part is conceptually of most interest, since it is close to the area of tissue breakdown. The fact that the paper points are inserted through the orifice might cause them to be saturated by fluids and microorganisms present in the upper part. Reaching the deeper part of the lesion, the paper points may have limited absorptive ability. Thus, a paper point sample might primarily reflect the microbiota of the orifice or the upper part of the lesion.
Baker and colleaguesCitation6 in an in vitro study used two microbial species and alternated placement of one of them in one of two separate layers in small wells. Paper points were inserted to the bottom of the wells and removed after 10 seconds. Irrespective of which bacteria were placed in the upper or lower layers, more than 90% of the total colony forming units recovered after culturing originated from the top layer. The authors suggested that this finding may be due to saturation of the paper points as they passed through the top layer.
In a study on periodontal lesions, Smola and colleaguesCitation7 compared samples taken by paper points with samples using foam tip swabs on the outer surface of the gingiva (‘pocket-out’ samples). The authors used polymerase chain reaction methods to detect five putative periodontal pathogens. No significant difference between the two sampling methods was detected for any of the pathogens. They suggested that both paper point and ‘pocket-out’ samples represented the microbiota present along the gingival margin.
In addition to sampling technique, the method to identify and quantify the putative periodontal pathogens may have an impact. To date, microbiological culturing has been the mainstay and is still considered to be the ‘gold standard’.Citation8–Citation10
The aim of this investigation was to study if positioning paper points to different depths during bacterial sampling of periodontal lesions has an impact on the recovery of three putative periodontal pathogens, using a culturing technique.
Material and methods
Subjects
Twenty patients, 30–60 years of age, to be treated at the Advanced Periodontics Clinic at Loma Linda University School of Dentistry were recruited. Records from initial periodontal examination were reviewed to identify patients having at least one periodontal lesion 6–8 mm deep with bleeding on probing at the mesio-buccal, mid-buccal, or disto-buccal aspect of a single-rooted tooth. Exclusion criteria consisted of patients: 1) having any systemic disease known to affect periodontal conditions; 2) having any condition for which antibiotic premedication was required; 3) having been on antibiotic treatment within the last three months; and 4) having had periodontal treatment within the last six months. Patients were enrolled in the study after signing an approved informed consent form between January and March 2006. Approval for the study was granted by the Institutional Review Board at Loma Linda University based upon the World Medical Association Declaration of Helsinki.
Clinical procedures
Following completion of the initial periodontal examination, volunteering and qualifying patients were given verbal and written information about the study and signed an informed consent. The patients returned for bacterial sample collection no sooner than one week later to ensure that the probing procedure would not distort the bacterial samples.
One examiner obtained all samples (author N.A.) using ISO 45 paper points (Dentsply, Tulsa, OK). The tooth to be sampled was isolated with cotton rolls and dried. The supragingival tooth surface was cleaned with a curette and subsequently with cotton rolls. Microbial samples were obtained by inserting points to three different depths of the periodontal lesions:
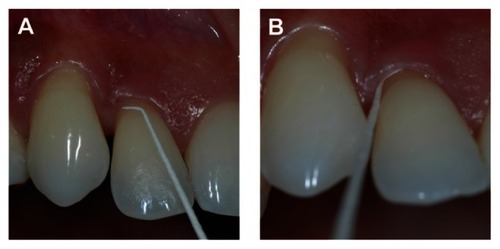
Orifice of lesion
The paper point was bent 2 mm from the tip of the point at an approximate 60° angle. The bent tip was placed horizontally along the gingival margin of the sampling site and inserted subgingivally until the bent tip was only slightly visible (). The insertion depth was thus related to the diameter of the paper tip at the 2 mm level and amounted to no more than 0.3–0.4 mm. The point was removed after 15 seconds, followed by retrieval of a second, similar orifice sample.
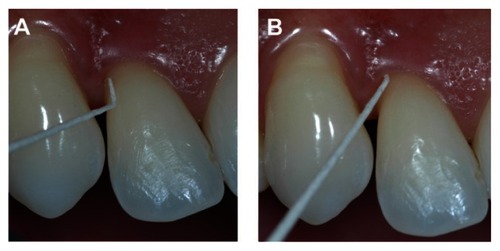
Two mm depth
Two paper points also bent 2 mm from the tip of the point were inserted at the same sampling site next to each other to this 2 mm depth and removed after 15 seconds ().
Base of lesion
Two paper points were inserted next to each other at the sampling site until resistance was met and removed after 15 seconds.
The sequence of sampling was always orifice level, followed by 2 mm level, followed by base level (an alternating/rotating sequence was not considered an option due to occurrence of bleeding following sampling the base of the lesion).
Microbiological identification methods
The two paper points from each sampling depth were pooled in 1 ml of anaerobic dental transport medium (Anaerobe Systems, Morgan Hill, CA) and stored at −20 °C until ready to use. The processing of the samples was performed in series of five patients at a time during the week following the sample collection (four weeks total).
For identification of Aggregatibacter actinomycetemcomitans, Brain Heart Infusion agar (Beckton Dickinson and Co, Sparks, MD) and broth with bacitracin and vancomycin (BHI/BV) were used. The BHI/BV agar/broth was prepared freshly every week by adding 75 mg/L bacitracin (Sigma Chemicals, St Louis, MO), 5 μg/L vancomycin (Sigma Chemicals), 6 mg/ml yeast extract (Beckton Dickinson and Co) and 0.1 ml/ml horse serum (Hyclone, Logan, UT).
For total anaerobic counts and identification of Porphyromonas gingivalis and Prevotella intermedia, Brain Heart Infusion agar (Beckton Dickinson and Co) and broth with 50 mg/L hemin (Sigma Chemicals), 15 μl/L vitamin K1 (Sigma Chemicals), 0.5 g/L l-cystein (Sigma Chemicals) and 45.5 ml/L sheep blood (Hemostat Laboratories, Dixon, CA) was also prepared fresh weekly (BHI/HK).
Prior to analyses of the paper point samples, strains of A. actinomycetemcomitans, P. gingivalis, and P. intermedia were purchased from American Type Culture Collection (ATCC, Manassas, VA) and cultured several times as described below to test the accuracy of the identification and enumeration of the colonies on the agar plates.
For the analyses of the paper point samples, 10-fold serial dilutions with the appropriate broth liquid were made from the samples following thawing and vortexing. One hundred μl of the various dilutions were dispersed onto the surface of agar plates with sterile L-shaped glass rods. The BHI/BV plates were incubated in 5% CO2 at 37 °C for 2–3 days. The BHI/HK plates were incubated in an anaerobic chamber (Coy Laboratory, Grass Lake, MI) at 37 °C for 5–7 days.
Counts of colony-forming units (CFUs) were performed on the plate dilution that resulted in 30–300 CFUs. If less than 30 CFUs were present at the lowest dilution plated, the number of colonies present was counted and recorded. However, the presence of a single colony was recorded as a 0 value.
For identification of A. actinomycetemcomitans, the plates were flooded with 3% H2O2 followed by counts of catalase positive colonies (presence of bubbles on colony) For identification of P. gingivalis and P. intermedia, longwave UV light (Fisher Scientific, Tustin, CA) and trypsin reaction were used. Dark-pigmented colonies that gave red fluorescence were identified as P. intermedia. Then, the plates were slightly misted with 500 mg/ml, pH 7.5 trypsin reagent (Sigma Chemicals) and re-examined under the UV light. Dark-pigmenting colonies with a blue fluorescence were identified as P. gingivalis.
Data analysis
The recovery of A. actinomycetemcomitans at the various levels of sampling was evaluated by comparison of the proportion of positive vs negative samples and by comparison of the CFU counts for positive samples. The recovery of P. gingivalis and P. intermedia was determined from calculations of the proportions of these microorganisms of the total anaerobic CFU counts. Non-parametric statistics using the Friedman test coupled with Bonferroni corrected post hoc comparisons were used to compare the CFU counts and the proportions of P. gingivalis and P. intermedia at the various levels of sampling.
Results
The recovery of the target microorganisms at the various levels of sampling the periodontal lesions is presented in . P. gingivalis and P. intermedia were recovered in all of the 20 patient samples for all the three testing depths. Out of the 20 samples, A. actinomycetemcomitans was found in eight orifice samples, eight 2 mm samples, and nine base samples. The concordance of positive samples comparing the different sampling levels was as follows: six sample sites were positive for both orifice and 2 mm level; five sample sites were positive for both orifice and base level; seven sample sites were positive for both 2 mm and base level; and five samples were positive for all three levels of sampling. The recovery for positive samples ranged between 0.01–5.0 CFUs × 103/ml.
Table 1 Presence of P. gingivalis, P. intermedia (proportion of positive samples) and A. actinomycetemcomitans (proportion of positive samples; CFUs × 103/ml for positive samples); total anaerobic CFUs × 105/ml; % P. gingivalis; and % P. intermedia in paper point samples taken at the orifice of the periodontal lesion, points inserted 2 mm into the lesion, and points inserted to the base of the lesion. Means ± SD N = 20
Total anaerobic CFUs × 105/ml ranged between 2.3–3.8 for the three sampling levels. Proportions of P. gingivalis ranged between 13.2%–21.2%. Proportions of P. intermedia ranged between 11.0%–16.4% (). The percentage of P. gingivalis for the 2 mm level samples was significantly different from the base samples. No other significant differences of recovery were observed.
Discussion
In the present study, A. actinomycetemcomitans was recovered from 40%–45% of the samples. This recovery rate is similar to that of previous studies using culturing.Citation11–Citation17
P. gingivalis and P. intermedia were recovered from all samples and in proportions ranging from 11% to 21%. Recovery of P. gingivalis was similar to that reported by some authors using culturing,Citation17–Citation20 but higher than that reported by other investigators.Citation13,Citation14,Citation16,Citation21–Citation23 Likewise, recovery of P. intermedia was similar to that reported by Gajardo and colleagues,Citation17 but higher than that reported by other investigators using culturing.Citation13–Citation16,Citation21–Citation23
The high recovery of P. gingivalis and P. intermedia in the present study may partly be explained by the fact that deep 6–8 mm lesions with bleeding on probing were sampled. The high recovery may also be explained by the identification method for these microorganisms in the present study. Identification was limited to test of fluorescence under long wave UV light and use of trypsin test. No additional confirmatory tests were employed. Nevertheless, this should not invalidate the findings of the present study, since the same methods were used to compare the recovery at the three different levels of sampling the lesions.
Although statistical difference was detected for the recovery of P.gingivalis from 2 mm depth of sampling, compared to the base of the lesion, the similarity in recovery of the target putative periodontal pathogens for the three different levels of sampling observed in the present study might be explained by two circumstances. As suggested by Baker and colleagues,Citation11 the paper points may become saturated as they pass through the orifice of the lesion and the absorption of microorganisms will therefore primarily occur at the orifice. This notion is supported by the findings by Smola and colleagues, Citation12 comparing paper point samples with samples using foam tip swabs applied on the outer surface of the gingiva from the gingival margin to the mucogingival junction (‘pocket-out’ samples). Based upon the similarity in recovery by the two sampling methods, they suggested that both paper point and ‘pocket-out’ samples may primarily represent the microbiota present along the gingival margin.
It is also conceivable that the pathogens may be present in similar proportions throughout the various depths of the periodontal lesions. If so, this seems to be contrary to the notion that the anaerobic putative periodontal pathogens primarily harbor the deeper parts of the lesions.Citation3,Citation24–Citation27
Conclusion
The findings of the present study seem to cast some doubts about the interpretation of microbial paper point sampling of periodontal lesions.
Disclosure
Author contributions: Nikola Angelov: patient selection, sample collection, culturing of bacterial samples; Raydolfo M Aprecio: culturing of bacterial samples, technical support; James Kettering: provided laboratory support, design of microbiological methods; Tord Lundgren: manuscript preparation, project support; Matt Riggs: statistical analysis; Jan Egelberg: project idea and design, manuscript preparation. The authors report no conflicts of interest in this work.
References
- ZambonJJPeriodontal diseases: microbial factorsAnn Periodontol199618799259118283
- HoltSCEbersoleJLPorphyromonas gingivalis, Treponema denticola and Tannerella forsythia: the “red complex”, a prototype polybacterial pathogenic consortium in periodontitisPeriodontol 20002005387212215853938
- SocranskySSHaffajeeADPeriodontal microbial ecologyPeriodontol 200020053813518715853940
- HartrothBSeyfahrtIConradsGSampling of periodontal pathogens by paper points: evaluation of basic parametersOral Microbiol Immunol19991432633010551161
- TannerACGoodsonJMSampling of microorganisms associated with periodontal diseaseOral Microbiol Immunol1986115223295677
- BakerPJButlerRWikesjoUMBacterial sampling by absorbent paper points. An in vitro studyJ Periodontol1991621421462027062
- SmolaSFRettenbergerGSimmetTBurysekLComparison of sample collection methods for the PCR detection of oral anaerobic pathogensLett Appl Microbiol20033610110512535130
- ZambonJJHaraszthyVIThe laboratory diagnosis of periodontal infectionsPeriodontol 20001995769829567931
- LoomerPMMicrobiological diagnostic testing in the treatment of periodontal diseasesPeriodontol 2000200434495614717855
- SanzMLauLHerreraDMorilloJMSilvaAMethods of detection of Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Tannerella forsythensis in periodontal microbiology, with special emphasis on advanced molecular techniques: a reviewJ Clin Periodontol2004311034104715560803
- ZambonJJChristerssonLASlotsJActinobacillus actinomycetemcomitans in human periodontal disease. Prevalence in patient groups and distribution of biotypes and serotypes within familiesJ Periodontol1983547077116358452
- RenvertSWikströmMDahlénGSlotsJEgelbergJEffect of root debridement on the elimination of Actinobacillus actinomycetemcomitans and Bacteroides gingivalis from periodontal pocketsJ Clin Periodontol1990173453502204636
- RodenburgJPvan WinkelhoffAJWinkelEGGoeneRJAbbasFde GraffJOccurrence of Bacteroides gingivalis, Bacteroides intermedius and Actinobacillus actinomycetemcomitans in severe periodontitis in relation to age and treatment historyJ Clin Periodontol1990173923992398137
- AliRWBakkenVNilsenRSkaugNComparative detection frequency of 6 putative periodontal pathogens in Sudanese and Norwegian adult periodontitis patientsJ Periodontol199465104610527853128
- Van der WeijdenGATimmermanMFReijerseEWolffeGNVan WinkelhoffAJVan der VeldenUThe prevalence of A. actinomycetemcomitans, P. gingivalis and P. intermedia in selected subjects with periodontitisJ Clin Periodontol1994215835887806673
- SalariMHKadkhodaZRate of cultivable subgingival periodontopathogenic bacteria in chronic periodontitisJ Oral Sci20044615716115508748
- GajardoMSilvaNGomezLPrevalence of periodontopathic bacteria in aggressive periodontitis patients in a Chilean populationJ Periodontol20057628929415974855
- LoosBClaffeyNCriggerMEffects of oral hygiene measures on clinical and microbiological parameters of periodontal diseaseJ Clin Periodontol1988152112163164329
- LoosBClaffeyNEgelbergJClinical and microbiological effects of root debridement in periodontal furcation pocketsJ Clin Periodontol1988154534633053787
- BoutagaKvan WinkelhoffAJVandenbroucke-GraulsCMSavelkoulPHComparison of real-time PCR and culture for detection of Porphyromonas gingivalis in subgingival plaque samplesJ Clin Microbiol2003414950495414605122
- SlotsJBacterial specificity in adult periodontitis. A summary of recent workJ Clin Periodontol1986139129173466908
- Jervoe-StormPMKoltzscherMFalkWDorflerAJepsenSComparison of culture and real-time PCR for detection and quantification of five putative periodontopathogenic bacteria in subgingival plaque samplesJ Clin Periodontol20053277878315966886
- RhemrevGETimmermanMFVeldkampIVan WinkelhoffAJVan der VeldenUImmediate effect of instrumentation on the subgingival microflora in deep inflamed pockets under strict plaque controlJ Clin Periodontol200633424816367855
- KigureTSaitoASeidaKYamadaSIshiharaKOkudaKDistribution of Porphyromonas gingivalis and Treponema denticola in human subgingival plaque at different pocket depths examined by immunohistochemical methodsJ Periodont Res1995303323417494175
- NoiriYShigeyukiEIdentification of periodontal disease-associated bacteria in the “plaque-free zone”J Periodontol2000711319132610972648
- NoiriYLiLEbisuSThe localization of periodontal disease-associated bacteria in human periodontal pocketsJ Dent Res2001801930193411706954
- NoiriYLiLYoshimuraFEbisuSLocalization of Porphyromonas gingivalis-carrying fimbriae in situ in human periodontal pocketsJ Dent Res20048394194515557402