Abstract
The development and in vitro performance of a modular nanoscale system capable of specific structural modification by enzymatic activity is described in this work. Due to its small physical size and adaptable characteristics, this system has the potential for utilization in targeted delivery systems and biosensing. Nanoparticle probes were synthesized containing two distinct fluorescent species including a quantum dot base particle and fluorescently labeled cleavable peptide substrate. Activity of these probes was monitored by gel electrophoresis with quantitative cleavage measurements made by fluorometric analysis. The model proximity-activated nanoparticles studied here exhibit significant susceptibility to cleavage by matrix metalloprotease-7 (MMP-7) at physiologically relevant concentrations, with nearly complete cleavage of available substrate molecules after 24 hours. This response is specific to MMP-7 enzyme activity, as cleavage is completely inhibited with the addition of EDTA. Utilization of enzyme-specific modification is a sensitive approach with broad applications for targeted therapeutics and biosensing. The versatility of this nanoparticle system is highlighted in its modular design, as it has the capability to integrate characteristics for detection, biosensing, targeting, and payload delivery into a single, multifunctional nanoparticle structure.
Introduction
Localized or specific delivery of therapeutics to diseased cells represents a historic paradigm shift in the treatment approach toward many disease states, including various types of cancer. Traditional approaches utilize systemic administration of highly potent drugs in an attempt to eradicate neoplastic cells, often with significant toxic side effects. Initial attempts at targeted delivery of these drugs have focused on the incorporation of small ligands to direct the therapy to a specific target site (CitationSachdeva 1998; CitationHallahan et al 2001; CitationPastorino et al 2003; CitationMitra et al 2005). However, the current generation of targeted therapies has not yet generally achieved highly specific localization to tumors and emerging neoplasia through a single recognition mechanism (CitationBogenrieder and Herlyn 2003; CitationKim and Toge 2004). Nonspecific binding and specific binding to nontarget cells compromise the therapeutic index of small molecule, ubiquitous cancer targeting ligands, and limits their applicability and efficiency as primary chemotherapeutic agents.
An alternative targeting approach involves the use of molecules capable of activation following cleavage by tumor- or other tissue-specific enzymes. These compounds have been incorporated into many structures ranging from in vivo imaging constructs (CitationWeissleder et al 1999; CitationTung et al 2000; CitationMcIntyre and Matrisian 2003; CitationHarris et al 2006) to chemically modified drug structures in the form of prodrugs (CitationSuzawa et al 2000, Citation2002; CitationLiu et al 2003). These studies demonstrate the feasibility of tumor-specific enzyme interaction as a means of modifying the physical structure of delivered drugs and imaging constructs, and potentially enables targeted delivery of drug carriers, including nanoparticles (NPs).
Several studies utilizing this approach have focused on the development of tumor-specific molecular imaging constructs cleavable by matrix metalloproteases (MMPs), including specific cleavable peptide sequences (CitationBremer et al 2001, Citation2002; CitationZucker and Cao 2001; CitationMcIntyre and Matrisian 2003; CitationMcIntyre et al 2004). The basis of the MMP-7 specific peptide substrate (RPLALWRS) utilized in one such report (CitationMcIntyre et al 2004) is employed in this study to impart tumor-specific activity to our NP proximity activated system.
The role of MMP-7 in biological processes
MMPs (including MMP-7) are a family of extracellular, zinc-dependent proteases that have a role in tissue breakdown and remodeling during both normal (eg, angiogenesis, gut innate immunity) and pathological (eg, inflammation, tumorigenesis, metastasis) processes. MMP-7 is important in the progression of a number of tumors, notably those originating in the colon or breast (CitationMcIntyre et al 2004). Furthermore, MMP-7 has proven to have diagnostic and prognostic value, as expression is associated with poor outcome in esophageal (CitationYamamoto et al 1999), colon (CitationAdachi et al 2001), and pancreatic (CitationYamamoto et al 2001) cancers.
For tumor systems in which MMP-7 activity is known to modulate the course of the disease, MMP-7 is secreted even in the earliest stages. Small, benign “precancerous” lesions, for example, exhibit MMP-7 secretion well before these lesions would be clinically detectable (CitationCrawford et al 2002; CitationHulboy et al 2004). Furthermore, analysis of MMP-7 expression in human neoplasia revealed that enzyme mRNA appeared in the surrounding normal tissue in only a few isolated cases (CitationWilson and Matrisian 1996), suggesting highly specific enzyme expression in the immediate tumor vicinity.
Nanoparticle-based therapeutic systems
Literature citations of tumor-specific cleavable constructs focus mainly on molecular imaging of enzyme activity and drug activation in vivo. However, incorporation of these constructs into NPs or their application to target NP-based therapeutic systems is an exciting area of opportunity. One recent study (CitationZhang et al 2006) has incorporated cleavable MMP substrates into NPs via a biotin-streptavidin linkage, outlining one method utilizing specific enzymatic activity to modulate NP uptake and delivery to cells. The work described here offers a conjugation approach that eliminates the need for biotin-streptavidin, resulting in a multifunctional nanoscale construct with novel characteristics. Importantly, studies described here include quantitative examination of NP construct cleavage and activation, providing a robust basis for further in vitro and in vivo targeting, delivery, and sensing applications.
Particulate systems have inherent advantages for these applications, in part, due to their relatively large surface to volume ratio, allowing the conjugation of multiple active molecules directly to the particle surface. Stable covalent conjugation of a range of bioactive molecules such as enzymes (CitationPhadtare et al 2004; CitationRossi et al 2004; CitationKuhn et al 2006, Citation2007), mAbs (CitationTang et al 2005), peptides (CitationLiu et al 2001), and DNA (CitationWang et al 2003; CitationZhang et al 2004) to the surface of NPs has been reported. Direct conjugation of ligands to the NP surface may overcome significant obstacles inherent in the ubiquitous biotin-streptavidin system, including immunogenicity, increased molecular size (primarily due to the relatively large molecular mass of streptavidin), and limited in vivo application. With small molecule ligands, many functional copies can potentially be directly attached to the surface of the NP enabling multiplexed interaction with target cells. In addition to these attractive nanoparticle characteristics, quantum dots (QDs) offer the added benefit of intense fluorescence characteristics for highly sensitive detection and imaging. The combination of these attributes impart the capability to integrate specific delivery, detection, imaging, and treatment characteristics into a single, multifunctional NP delivery vehicle.
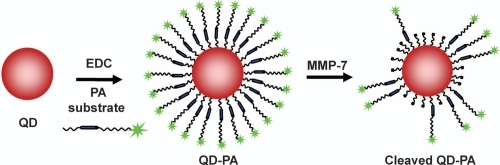
In this study, we describe the synthesis and in vitro performance of a NP probe capable of specific activation by MMP-7. In addition, the importance of quantitative analysis of construct performance is described in the context of in vitro and in vivo application. To construct the proximity activated NP, a carboxylated QD was functionalized with a peptide construct (PEG3400-[Ahx]-RPLALWRS-[Ahx]-PEG5000-K(5-FAM); PA construct). The PA construct consists of an 8-mer peptide substrate (RPLALWRS) cleavable by MMP-7. The PA construct also contains polyethylene glycol (PEG) groups to reduce nonspecific binding. Therefore, the PA construct initially presents as a PEGylated nanoparticle with low nonspecific binding as predicted from previous studies (CitationAkerman et al 2002; CitationBallou et al 2004; CitationGao et al 2004). Cleavage of the internal peptide liberates the distal PEG and part of the peptide, along with a fluorophore, in this work, for assessment of cleavage (). In the future, proteolytic modification could be used to “unmask” the particle, enabling access to underlying small molecule ligands previously concealed near the nanoparticle surface. Effective PA targeting will likely be modulated by many molecular factors, including the relative characteristics of the distal and proximal PEGs on the PA construct and the PEG (or other molecular spacer) that displays the ligand from the nanoparticle surface.
Materials and methods
Cleavable peptides and NP probes
The PEGylated substrate for MMP-7 (PEG3400-[Ahx]-RPLALWRS-[Ahx]-PEG5000-K(5-FAM; PA construct) was obtained from Anaspec. This structure consists of a cleavable 8-mer peptide substrate (RPLALWRS) flanked by polyethylene glycol (PEG) groups of 3400 and 5000 g/gmol and aminohexanoic acid (Ahx) groups to serve as spacer molecules, as well as a fluorescent tag (5-FAM) for cleavage detection. The N-terminus of the construct was a primary amine to facilitate further conjugation reactions. To stabilize the substrate and prevent additional modification, C-terminal amidation was performed during synthesis.
The N-terminal amine group of the substrate was conjugated to the surface of carboxylate QDs (CdSe dots coated with ZnS, 585 nm emission; Quantum Dot Corp., Hayward, CA) using 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC). 250 μl of QDs (7.5 μM) was diluted to 1 μM with 10 mM borate buffer, pH 7.4. 80 μL from a 10 mg/mL stock solution of the cleavable PA substrate was added to the solution, yielding a PA:QD molar ratio of 10.0:1. 57 μl from a 10 mg/ml stock solution of EDC was added and the mixture was stirred for 2 h at room temperature to complete the conjugation. Unconjugated PA construct was removed by performing three buffer exchanges in a 50,000 Da nominal molecular weight limit (NMWL) cutoff spin filter device (Amicon Ultra). The concentration of the final purified product was determined by measuring the absorbance at 575 nm using a QD extinction coefficient of 400,000 M−1cm−1.
MMP-7 activity assay
Studies were conducted to determine the susceptibility of both the PA and QD-PA constructs to exogenous MMP-7. Active MMP-7 (Calbiochem) was added to 100 μl of a 50 nM PA or QD-PA solution in borate buffer (pH 7.4) supplemented with 50 μM ZnSO4. The total concentration of MMP-7 was varied in these samples from 100 nM to 5 nM. Samples were then incubated at 37 °C for various timepoints ranging from 1 h to 48 h. Control studies involving QD-PA with no enzyme and with inactivated enzyme (by addition of 30 mM EDTA) were carried out simultaneously for direct comparison. In the case of QD-PA constructs, any cleavage products were removed by spin filtration in a 50,000 NMWL cutoff device.
Gel electrophoresis
PA constructs were analyzed on a 10%–20% gradient tris-tricine polyacrylamide gel (Life Gels). Samples were loaded in a buffer containing 10% glycerol in tris-tricine running buffer (Life Gels). Gels were then run under a 150 mV voltage for ~1 h. Control, unconjugated PA constructs were compared with cleaved PA constructs to straightforwardly visualize and confirm enzymatic activity against the substrate.
A quantitative determination of PA construct cleavage was made by digital analysis of gel electrophoresis images. Gel images were obtained as JPEG files and visually analyzed in Doc-It LS software (UVP Inc.). Regions of interest were drawn around individual bands, and each band was electronically cut from the image and saved as a separate JPEG file. These files were loaded into a simple MATLab script designed to calculate quantitative image parameters. The composite fluorescence intensity for each band was calculated by correcting for background intensity and then summing the measured intensity of each pixel contained in the image. The relative percentage contained in each band was obtained by comparing its composite fluorescence to that of the total for the lane.
QD-PA constructs were assayed by loading samples directly on to a 0.8% bufferless agarose gel (E-Gel, Invitrogen Corp.). QDs subjected to the cleavage reaction were assayed against native carboxylate QDs, uncleaved QD-PA constructs, and QD-PA + EDTA. All gels were imaged for fluorescence on a BioDoc-It imaging system (UVP Inc.).
Fluorescence spectroscopy
Following PA conjugation and purification, the fluorescence spectra of QD-PA constructs were measured by fluorescence spectroscopy at 488 nm excitation (Perkin-Elmer LS 50B). Enzymatic cleavage of the QD-PA constructs was monitored by measuring changes to the fluorescence spectra following incubation with MMP-7. The spectra were scaled to normalize QD peak fluorescence (~585 nm) between all samples. The peak fluorescence intensity values at ~519 nm and ~585 nm were then directly compared to the spectra of control, uncleaved QD-PA particles to calculate the extent of PA construct cleavage following MMP-7 treatment.
Size measurements
Nanoparticle size was determined using dynamic light scattering (DLS) measurements. DLS measurements were conducted on a Malvern Nano Series Zetasizer with a 633 nm laser. The duration of the scans was 60 s, and 3 scans were accumulated. The concentration of all QD samples was 0.25 μM.
Statistics
Statistical significance of experimental populations was assessed using an upaired t-test and determined by Sigma-Stat (Jandel Scientific Software). Differences were termed significant for p < 0.05.
Results and discussion
In vitro cleavage of the PA construct
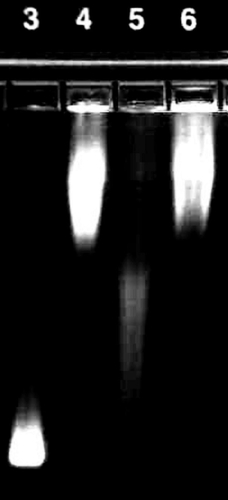
The PA construct is highly susceptible to cleavage by MMP-7. Prior to conjugation to the QD surface, PA constructs were assayed for susceptibility to enzymatic activity. Analysis of gel electrophoresis images of unconjugated PA (prior to conjugation to the NP surface) confirms specific cleavage of these constructs by MMP-7 (). Lanes 1 and 2 confirm the presence of the intact, fluorescently labeled PA structure. These bright, low mobility bands are nearly extinguished after 24 h exposure to MMP-7. This cleavage also results in the generation of a very bright, high mobility band corresponding to the low molecular weight, labeled cleavage product (, lanes 3 and 4). PA construct cleavage was completely abolished by the addition of 30 mM EDTA (a zinc chelator; , lanes 5 and 6), as the band corresponding to the cleaved PA construct is absent in these wells. These results are consistent with the near complete proteolysis of the PA construct at the site of the MMP target peptide and inhibition of the zinc-dependent MMP-7 enzyme by EDTA.
Figure 2 Gel electrophoresis confirms cleavage of the soluble, unconjugated PA construct by MMP-7. Lanes 1 and 2 show the unconjugated PA construct prior to enzymatic exposure as a control. The bright, low mobility band (identified by the white arrow) represents the intact high molecular weight PA structure. Lanes 3 and 4 represent the PA construct exposed to MMP-7 for 24 hours, while lanes 5 and 6 show the inhibition of MMP-7 activity by EDTA.
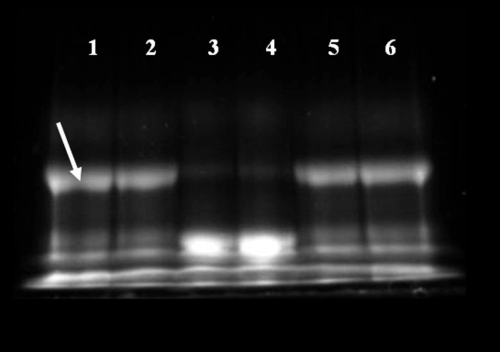
Figure 3 The surface of carboxylate QDs was successfully functionalized with the cleavable PA construct. Mobility of QDs modified with the cleavable PA construct (lane 4, diffuse band) is reduced compared with that of unmodified QDs (lane 3). 24 h exposure of the QD-PA conjugates to MMP-7 (lane 5) leads to a partial recovery of mobility, Increased mobility after exposure to MMP-7 can be completely inhibited by the addition of 30 mM EDTA (lane 6). This image is representative of three independent trials.
A quantitative determination of PA construct cleavage was made by digital analysis of gel electrophoresis images. From this analysis, the percentage of PA construct contained in each sample was determined. Control, uncleaved soluble PA construct contains 35.6 ± 3.4% intact PA structure, as determined from 4 independent trials. The remaining 64.4% of the fluorescence intensity in these bands represents high mobility structures consistent with 5-FAM labeled reactants and intermediates. 4.6 ± 0.7% of the intact PA structure remains after exposure to 100 nM MMP-7 for 24 hours. Therefore, 87.2 ± 0.7% of the initial intact PA construct was cleaved (p < 0.001) and 12.8 ± 0.7% remained intact after enzymatic exposure, confirming the high susceptibility of the PA construct to MMP-7 proteolysis. Samples treated with MMP-7 + 30 mM EDTA contain 35.5 ± 2.0% intact PA structure, a value equivalent to the amount measured in control samples in the absence of MMP-7 (p = 0.966).
QD-PA construct synthesis and activity
The surface of carboxylate QDs was successfully modified with the PA construct. This was evident as the size of the nanoparticle increased after functionalization. The size of the unmodified QD is 18.2 ± 0.3 nm, according to dynamic light scattering (DLS) measurements. After functionalization, the size of the quantum dot functionalized with the PA construct (QD-PA) is 81.8 ± 10.5 nm. Additionally, agarose gel electrophoresis shown in reveals that conjugation of the PA construct to the QD surface results in a significant decrease in electrophoretic mobility (lane 4) over unmodified carboxylate QDs (lane 3), presumably corresponding to an increase in MW and surface charge characteristics of the QD following PA conjugation. MMP-7 treatment results in partial recovery of mobility (lane 5), consistent with a reduction of surface-associated molecular mass mediated by proteolytic cleavage. Mobility recovery is not complete, consistent with persistence of the proximal portion of the PA construct on the QD surface after proteolytic cleavage. MMP-7 mediated increase in mobility is reversed with the addition of 30 mM EDTA during cleavage (lane 6), suggesting inhibition of MMP-7 activity by EDTA. Changes in relative mobilities provide evidence of successful QD surface functionalization, subsequent proteolytic degradation, and inhibition of enzyme activity expected in this system.
QD-PA synthesis and activity was further confirmed by measuring the fluorescence spectra of the resulting conjugates. Following conjugation and purification, the fluorescence spectra of the QD-PA constructs were measured (). These spectra reveal fluorescence maxima at approximately 520 and 585 nm, corresponding to the emission maxima of the 5-FAM labeled PA construct (519 nm) and of the QD base (585 nm), respectively.
Figure 4 QDs modified with PA construct possess two fluorescence emission peaks consistent with colocalized QD and PA construct fluorescence. Exposure to MMP-7 (dashed line) results in reduced 5-FAM fluorescence intensity at approximately 520 nm when compared to control, uncleaved QD-PA samples (solid line). Results shown are representative of n ≥ 3 trials for all experimental groups.
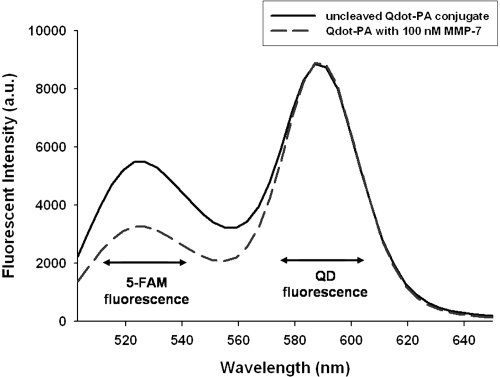
Enzymatic cleavage was monitored by measuring changes to the fluorescence spectra of the QD-PA constructs following incubation with MMP-7 (, QD-PA + 100 nM MMP-7). Relative to control, uncleaved QD-PA constructs, these spectra reveal a reduced fluorescence peak at 519 nm, suggesting loss of 5-FAM fluorescence from the QD-PA construct, consistent with proteolysis of the PA construct at the site of the MMP-7 target peptide and subsequent removal of cleavage products. This result is consistent with gel electrophoresis images and computed results, demonstrating retention of PA proteolytic susceptibility following conjugation to the QD surface. Additional studies revealed no significant photobleaching of either 5-FAM or QD over the course of the measurements. Furthermore, no significant change in QD fluorescence intensity (at 585 nm) was observed upon PA cleavage from the surface.
Fluorescence measurements were used to produce a quantitative analysis of PA construct cleavage. These measurements () confirm significant cleavage of the PA construct from the QD surface. Cleavage increased steadily with incubation time, reaching a plateau after 24 h (90.9 ± 15.4% cleavage), consistent with cleavage of unconjugated PA constructs under the same conditions (87.1 ± 0.7% cleavage). Measured cleavage values are highly dependent on incubation time, following an exponential rise to maximum curve describing traditional enzyme kinetics.
Figure 5 Fluorescence spectra of control, uncleaved QD-PA structures were compared to those of samples incubated with variable concentrations of MMP-7. Statistical measurements: * = p < 0.05, **p < 0.001. n ≥ 3 for all concentrations and timepoints.
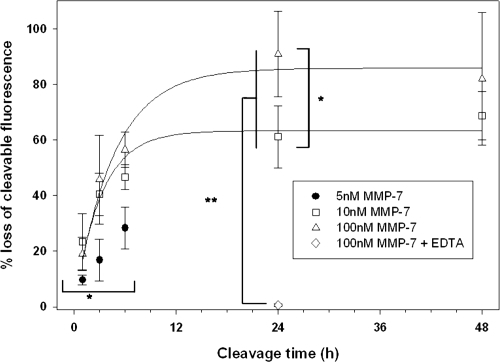
Cleavage of the PA construct from the QD surface was also influenced by initial MMP-7 concentration. Cleavage was measured from samples using MMP-7 concentrations ranging from 100 nM to 5 nM (reflecting the range of expected concentrations in vivo) as shown in . Although the cleavage response measured for each MMP-7 concentration follows the same general trend, the extent of cleavage is markedly altered, declining as MMP-7 concentration was reduced. Cleavage at early timepoints (t ≤ 6 h) is insensitive to MMP-7 concentrations above approximately 10 nM.
Figure 6 Cleavage of PA construct from the QD surface at early timepoints (t < 6 h) is modulated by MMP-7 concentration. Quantitative cleavage measurements were analyzed in the form of a dose response curve to highlight the influence of enzyme concentration on construct cleavage. * = p < 0.05, n ≥ 3 for all MMP-7 concentrations and timepoints.
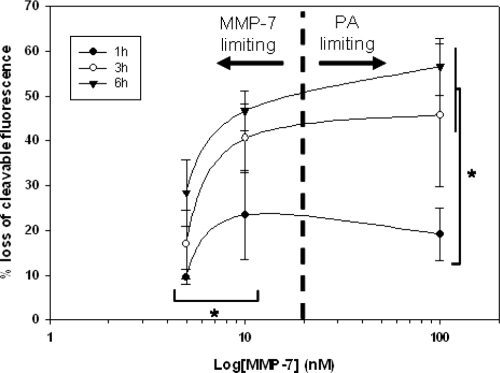
Previous literature reports the Km of unmodified PA substrate as 26 μM (CitationWelch et al 1995). Thus, at substrate concentrations below this value, PA cleavage is expected to be independent of MMP-7 concentration. Bulk PA concentrations used in the current study range from 37.5–100 nM, well below the reported Km. However, at the given substrate concentration, and at low MMP-7 concentrations (eg, 5 nM and 10 nM), MMP-7 is the limiting reagent. At higher MMP-7 concentrations (eg, 100 nM), MMP-7 concentration meets or exceeds substrate concentration. Thus, cleavage behavior transitions from a regime where MMP-7 is the limiting component to a regime where PA is the limiting component, resulting in measurements consistent with conventional enzyme kinetics.
Literature has shown that MMP-7 concentration can vary widely, with overexpression noted in various tumor phenotypes (CitationMcIntyre et al 2004). While few quantitative measures of MMP-7 concentration have been performed, previous studies have utilized concentrations of up to 100 nM in vitro (CitationKioi et al 2003; CitationWang et al 2005), while others have measured comparable concentrations of additional MMPs in vivo (CitationFuruya et al 2000). This evidence suggests likely local MMP-7 concentrations in tumor of at least 50 nM, corresponding with the levels utilized in the current study. Furthermore, our studies reveal that proteolytic susceptibility of QD-PA probes saturates at MMP-7 concentrations above 10 nM. Thus, at enzyme concentrations at or above this threshold, QD-PA probes are expected to exhibit maximal activity.
As particle activation time and tissue residence time (ie, particle velocity) are competing factors, this ratio is an important consideration with potential for significant impact on in vivo utility of QD-PA constructs. If activation time is long compared to residence time (eg, for vascular targets), then the NP will presumably be transported away during activation and before specific interaction can occur. In this case, PA delivery has characteristics similar to those for current targeted approaches. However, the small size and surface passivation (PEGylation) of the QD-PA construct combine to create a construct likely capable of extravasation and tumor accumulation, enabling access to targets within the tissue. In this compartment, particle transport is typically very slow, allowing ample time for particle activation. At the predicted tissue MMP-7 concentrations, QD-PA probes require <6 h for activation (defined as 50% cleavage), with significant activity reached within 3 h. This activation time relates well to NP tissue residence times (CitationKaul and Amiji 2004; CitationMontet et al 2006), yielding probe performance characteristics consistent with in vivo triggered targeting.
Conclusions
The QD-PA constructs described here could potentially provide a powerful, versatile platform for targeted delivery and biosensing for both in vitro and in vivo applications. Although the importance of therapeutic targeting has been acknowledged for more than two decades, few viable options for targeted therapy have been successfully implemented. Many treatment regimens continue to rely on systemic administration that compromises drug concentrations delivered to target tissue based on toxicity resulting from specific and nonspecific delivery to nontarget cells (CitationFrei 1997).
The modular structure of the QD-PA constructs provides multiple parameters for performance optimization, with various factors presumably modulating the overall activity of the construct. Successful optimization requires consideration of the nanoscale, biological chemistry, and physiological transport of the overall construct. PEG length is one variable with potential to significantly affect multiple characteristics including enzyme susceptibility (by limiting enzyme access to the substrate), nonspecific binding limitation, effective ligand concealment (in future structures), and particle mobility. Additional structural parameters including enzyme substrate configuration, ligand selection, and QD ligand surface density (number of molecules conjugated to the QD surface) can also directly affect probe performance. Tailoring of these multiple characteristics should enable optimization of probe performance for the requirements of both in vitro and in vivo systems.
PA targeting requires the colocalization of the cleavable mask and the targeting ligand, necessitating a particulate approach. In the exemplar described here, the NP is a QD, used to provide a fluorescent beacon for performance assessment. Subsequent work will employ this construct, amended to include a targeting ligand, to assess proteolytically activated targeting in vitro and in vivo by quantitative fluorescence measurements and fluorescence imaging. While the appropriateness of QDs for clinical applications remains theoretically limited due to their heavy metal content, the PA targeting approach described here can be applied to other nanostructures approved for use in humans. These nanomaterials include iron oxide nanoparticles for magnetic resonance image contrast enhancement and drug delivery structures such as polymers, liposomes, polymersomes, and gene delivery vehicles.
Acknowledgements
The authors gratefully acknowledge the Department of Defense Idea Award BC043908 for funding.
References
- AdachiYYamamotoHItohFClinicopathologic and prognostic significance of matrilysin expression at the invasive front in human colorectal cancersInt J Cancer200195290411494227
- AkermanMEChanWCLaakkonenPNanocrystal targeting in vivoProc Natl Acad Sci USA200299126172112235356
- BallouBLagerholmBCErnstLANoninvasive imaging of quantum dots in miceBioconjug Chem200415798614733586
- BogenriederTHerlynMAxis of evil: molecular mechanisms of cancer metastasisOncogene20032265243614528277
- BremerCTungCHWeisslederRIn vivo molecular target assessment of matrix metalloproteinase inhibitionNat Med20017743811385514
- BremerCTungCHWeisslederRMolecular imaging of MMP expression and therapeutic MMP inhibitionAcad Radiol20029Suppl 2S3141512188259
- CrawfordHCScogginsCRWashingtonMKMatrix metalloproteinase-7 is expressed by pancreatic cancer precursors and regulates acinar-to-ductal metaplasia in exocrine pancreasJ Clin Invest200210914374412045257
- FreiEIIIAntmanKHHollandJFFreiEIIIBastRCCombination chemotherapy, dose and scheduleCancer Medicine19974PhiladelphiaWilliams and Wilkins81737
- FuruyaMIshikuraHOgawaYAnalyses of matrix metalloproteinases and their inhibitors in cyst fluid of serous ovarian tumorsPathobiology2000682394411493755
- GaoXCuiYLevensonRMIn vivo cancer targeting and imaging with semiconductor quantum dotsNat Biotechnol2004229697615258594
- HallahanDEGengLCmelakAJTargeting drug delivery to radiation-induced neoantigens in tumor microvasculatureJ Control Release2001741–31839111489494
- HarrisTJvon MaltzahnGDerfusAMProteolytic actuation of nanoparticle self-assemblyAngew Chem Int Ed20064531615
- HulboyDLGautamSFingletonBThe influence of matrix metalloproteinase-7 on early mammary tumorigenesis in the multiple intestinal neoplasia mouseOncol Rep200412131715201952
- KaulGAmijiMBiodistribution and targeting potential of poly(ethylene glycol)-modified gelatin nanoparticles in subcutaneous murine tumor modelJ Drug Target2004129–105859115621684
- KimRTogeTChanges in therapy for solid tumors: potential for overcoming drug resistance in vivo with molecular targeting agentsSurg Today20043429330315052442
- KioiMYamamotoKHigashiSMatrilysin (MMP-7) induces homotypic adhesion of human colon cancer cells and enhances their metastatic potential in nude mouse modelOncogene20032286627014647460
- KuhnSJFinchSKHallahanDEProteolytic surface functionalization enhances in vitro magnetic nanoparticle mobility through extracellular matrixNano Lett200663061216464055
- KuhnSJFinchSKHallahanDEFacile production of multivalent enzyme-nanoparticle conjugatesJournal of Magnetism and Magnetic Materials20073116872
- LiuCSunCHuangHOverexpression of legumain in tumors is significant for invasion/metastasis and a candidate enzymatic target for prodrug therapyCancer Res20036329576412782603
- LiuJZhangQRemsenEENanostructured materials designed for cell binding and transductionBiomacromolecules20012362811749193
- McIntyreJOFingletonBWellsKSDevelopment of a novel fluorogenic proteolytic beacon for in vivo detection and imaging of tumour-associated matrix metalloproteinase-7 activityBiochem J2004377Pt 36172814556651
- McIntyreJOMatrisianLMMolecular imaging of proteolytic activity in cancerJ Cell Biochem20039010879714635184
- MitraAMulhollandJNanATargeting tumor angiogenic vasculature using polymer-RGD conjugatesJ Control Release200510219120115653145
- MontetXMontet-AbouKReynoldsFNanoparticle imaging of integrins on tumor cellsNeoplasia200682142216611415
- PastorinoFBrignoleCMarimpietriDVascular damage and anti-angiogenic effects of tumor vessel-targeted liposomal chemotherapyCancer Res2003637400914612539
- PhadtareSVinodVPMukhopadhyayKImmobilization and biocatalytic activity of fungal protease on gold nanoparticle-loaded zeolite microspheresBiotechnol Bioeng2004856293714966804
- RossiLMQuachADRosenzweigZGlucose oxidase-magnetite nanoparticle bioconjugate for glucose sensingAnal Bioanal Chem20043806061315448967
- SachdevaMSDrug targeting systems for cancer chemotherapyExpert Opin Investig Drugs19987184964
- SuzawaTNagamuraSSaitoHEnhanced tumor cell selectivity of adriamycin-monoclonal antibody conjugate via a poly(ethylene glycol)-based cleavable linkerJ Control Release2002792294211853934
- SuzawaTNagamuraSSaitoHSynthesis and HPLC analysis of enzymatically cleavable linker consisting of poly(ethylene glycol) and dipeptide for the development of immunoconjugateJ Control Release200069274111018544
- TangDYuanRChaiYNovel potentiometric immunosensor for determination of diphtheria antigen based on compound nanoparticles and bilayer two-dimensional sol-gel as matricesAnal Bioanal Chem20053816748015657709
- TungCHMahmoodUBredowSIn vivo imaging of proteolytic enzyme activity using a novel molecular reporterCancer Res2000604953810987312
- WangFQSoJReierstadSMatrilysin (MMP-7) promotes invasion of ovarian cancer cells by activation of progelatinaseInt J Cancer2005114193115523695
- WangMSunCWangLElectrochemical detection of DNA immobilized on gold colloid particles modified self-assembled monolayer electrode with silver nanoparticle labelJ Pharm Biomed Anal20033311172514656602
- WeisslederRTungCHMahmoodUIn vivo imaging of tumors with protease-activated near-infrared fluorescent probesNat Biotechnol199917375810207887
- WelchARHolmanCMBrownerMFPurification of human matrilysin produced in Escherichia coli and characterization using a new optimized fluorogenic peptide substrateArch Biochem Biophys199532459647503560
- WilsonCLMatrisianLMMatrilysin: an epithelial matrix metalloproteinase with potentially novel functionsInt J Biochem Cell Biol199628123368729000
- YamamotoHAdachiYItohFAssociation of matrilysin expression with recurrence and poor prognosis in human esophageal squamous cell carcinomaCancer Res19995933131610416584
- YamamotoHItohFIkuSExpression of matrix metalloproteinases and tissue inhibitors of metalloproteinases in human pancreatic adenocarcinomas: clinicopathologic and prognostic significance of matrilysin expressionJ Clin Oncol20011911182711181677
- ZhangDChenYChenHYSilica-nanoparticle-based interface for the enhanced immobilization and sequence-specific detection of DNAAnal Bioanal Chem20043797–810253015179534
- ZhangYSoMKRaoJProtease-modulated cellular uptake of quantum dotsNano Lett2006619889216968013
- ZuckerSCaoJImaging metalloproteinase activity in vivoNat Med20017655611385494