Abstract
Nowadays diabetic nephropathy (DN) is the most common cause of end-stage renal disease (ESRD). Recent studies have demonstrated that the myosin, heavy chain 9, non-muscle (MYH9) gene is associated with ESRD in African Americans. In this study, we tested the hypothesis that a common single nucleotide polymorphism rs16996677 in the MYH9 gene may contribute to the etiology of DN in type 2 diabetes (T2D) in a Taiwanese population with T2D. There were 180 T2D patients diagnosed with DN and 178 age- and sex-similar T2D without DN controls. Single locus analyses showed no significant main effects of MYH9 rs16996677 on the risk of DN in T2D. The results suggest that the rs16996677 SNP in MYH9 may not contribute to the risk of DN in T2D in Taiwanese T2D patients.
Introduction
End-stage renal disease (ESRD) is the fifth stage of chronic kidney disease when the kidneys permanently fail to work.Citation1,Citation2 The number of ESRD patients treated through dialysis or transplantation has continued to grow worldwide.Citation1,Citation2 Nowadays diabetic nephropathy (DN) is the leading reported cause of ESRD.Citation1,Citation2 DN is a common microvascular complication of diabetes (for type 1 and type 2),Citation3 and type 2 diabetes (T2D) is characterized by insulin insensitivity and pancreatic beta-cell dysfunction.Citation4,Citation5 More and more genetic variants associated with ESRD are being discovered using candidate gene approaches, family linkage studies, gene expression profiling, and genome-wide association studies.Citation6–Citation8 The identification of genetic variants that predispose to ESRD will enhance our understanding of the pathophysiology of renal disorders, thereby potentially leading to novel tailored therapies for treatment and prevention.Citation7,Citation8
The myosin, heavy chain 9, non-muscle (MYH9) gene encodes the protein non-muscle myosin heavy chain, class II, and isoform type A, which is abundantly expressed in the kidney, liver, and platelets.Citation9,Citation10 Recently, the MYH9 gene has received much attention. Some studies have suggested that the MYH9 gene is associated with non-diabetic ESRD,Citation11 hypertension-associated ESRD,Citation12,Citation13 and T2D-associated ESRDCitation14 in African Americans. In addition, MYH9 has been linked with idiopathic and human immunodeficiency virus-associated focal segmental glomerulosclerosis in African Americans.Citation12 It has also been shown that there is an association between MYH9 and albuminuria was in hypertensive African Americans.Citation15 Moreover, a common single nucleotide polymorphism (SNP) rs16996677 in MYH9 has been found to be highly significantly associated with non-diabetic ESRD,Citation11 hypertension-associated ESRD,Citation13 and T2D-associated ESRDCitation14 in a population of African Americans. Furthermore, it has been reported that MYH9 rs16996677 was not associated with kidney failure by contrasting the T2D-associated ESRD cases with T2D lacking nephropathy controls.Citation14
The previous findingsCitation11,Citation13,Citation14 mainly reported of the association studies of the MYH9 rs16996677 polymorphism and ESRD in African Americans. In this work, we focused on T2D-associated ESRD due to DN and tested the hypothesis that the rs16996677 polymorphism in the MYH9 gene may contribute to the etiology of DN in T2D amongst Taiwanese T2D individuals.
Materials and methods
Patients
The patients were partially original to the previous study by Wu et alCitation16 and are described in detail elsewhere.Citation16 Briefly, there were 358 Taiwanese patients with T2D who were recruited from the Tri-Service General Hospital in Taipei, Taiwan, in 2002. The case group comprised 180 T2D patients with DN and the control group comprised 178 T2D patients without DN. All the recruited patients fulfilled the following four inclusion criteria: (1) The patient had been diagnosed with diabetes for more than 5 years; (2) The age was between 30 and 75 years; (3) The fasting plasma glucose was greater than 126 mg/dl; (4) The glycated haemoglobin was greater than 6%. We then further classified the study subjects as DN or T2D without DN groups according to three surrogate endpoints, comprising urinary albumin to creatinine ratio, blood urea nitrogen, and serum creatinine.
Before conducting the study, approval was obtained from the Internal Review Board of the Tri-Service General Hospital and the approved informed consent form was signed by each subject.
Laboratory methods
provides detailed information on the selected SNP, which includes its chromosome position, commercial assay identifier, allelic variants, and the minor allele frequency. DNA was isolated from blood samples using a QIAamp
Table 1 Chromosome position, commercial assay identifier, nucleotide variation, and minor allele frequency of the selected SNP
DNA blood kit following the manufacturer’s instructions (Qiagen, Valencia, CA). To extract DNA, we used 200 μl of blood which was further solved in 200 μL of distilled water.Citation16 Before PCR reaction, part of the extracted DNA was diluted into a concentration of 10 μg/μL. The qualities of isolated genomic DNAs were checked using agarose gel electrophoresis and the quantities determined using spectrophotometry.
All SNP genotypings were performed using the Taqman SNP genotyping assay (Applied Biosystems Inc. Foster City, CA). The primers and probes of SNPs were from the ABI Assay on Demand kit. Reactions were carried out according to the manufacturer’s protocol. The probe fluorescence signal detection was performed using the ABI Prism 7900 Real-Time PCR system.
HapMap database
In this work, we utilized the HapMap databaseCitation17 to provide a comparison between a Taiwanese population and the five populations (African ancestry in Southwest USA [ASW], Utah residents with Northern and Western European ancestry from the CEPH collection [CEU]; Han Chinese in Beijing, China [CHB]; Japanese in Tokyo, Japan [JPT]; Yoruba in Ibadan, Nigeria [YRI]) of the HapMap database in terms of allele frequencies.
Statistical analysis
The categorical data were analyzed using the chi-square test. Furthermore, we compared differences for continuous variables using the Student’s t-test. In addition, genotype frequencies were evaluated for Hardy-Weinberg equilibrium using a χ2 goodness-of-fit test. The criterion for significance was set at P < 0.05 for all tests. Data are presented as mean ± standard deviation.
Results
describes the demographic and clinical characteristics of the study population. As shown in , unrelated DN cases and T2D without DN controls had a similar gender distribution (P = 0.1116). In addition, the distribution of age in the two groups was well-matched (P = 0.1008). All SNPs were evaluated for their contribution to DN in the complete sample population, including 180 DN subjects and 178 T2D without DN subjects.
Table 2 Demographic and clinical characteristics of study subjects
shows the genotype and allele distributions of the rs16996677 SNP in the DN (case) and T2D without DN (control) groups for the complete sample population. No association between MYH9 rs16996677 and DN was detected by the allelic or genotypic test (P = 0.3156 and 0.3153, respectively). In addition, genotype frequency distribution for rs16996677 was in Hardy-Weinberg equilibrium (; P = 0.9788).
Table 3 Distributions of genotypes and alleles between the diabetic nephropathy (DN) cases and type 2 diabetes (T2D) without DN controls
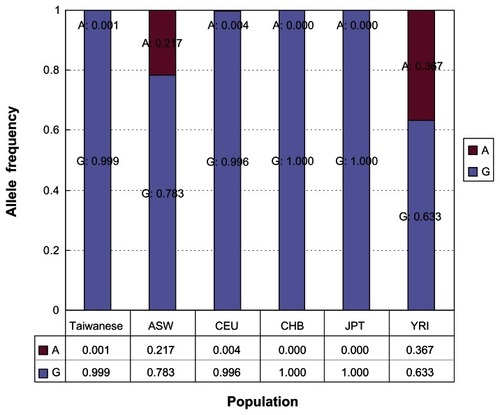
Furthermore, demonstrates the allele frequencies of the rs16996677 SNP in the MYH9 gene for the five populations of the HapMap database and the Taiwanese population collected in this study. We compared the allele frequencies between the Taiwanese population and each of the five populations of HapMap (that is, Taiwanese versus ASW, Taiwanese versus CEU, Taiwanese versus CHB, Taiwanese versus JPT, and Taiwanese versus YRI). There was a significant difference between Taiwanese and ASW populations (P < 0.0001) as well as between Taiwanese and YRI populations (P < 0.0001). However, we did not find a significant difference between Taiwanese and CEU populations (P = 0.3883), between Taiwanese and CHB populations (P = 0.7216), or between Taiwanese and JPT populations (P = 0.7216).
Figure 1 The allele frequencies of the rs16996677 SNP in the MYH9 gene for the five populations of the HapMap database and the Taiwanese population collected in this study.
Discussion
Our study is the first to date to have examined whether the MYH9 rs16996677 polymorphism is associated with the risk of DN amongst Taiwanese T2D individuals. In this study, MYH9 rs16996677 was not associated with DN in T2D (in either allelic or genotypic tests) using single locus analyses. Similarly, Freedman et al compared the T2D-associated ESRD patients with T2D lacking nephropathy controls and found no association between MYH9 rs16996677 and kidney failure among African Americans.Citation14 In contrast, Freedman et al further reported a significant association between MYH9 rs16996677 and T2D-associated ESRD by comparing T2D-associated ESRD cases with non-diabetic non-nephropathy controls in the same African American study.Citation14 In a previous Taiwanese study, Wu et al also suggested that the SNP rs1044498 in the ectonucleotide pyrophosphatase/phosphodiesterase 1 (ENPP1) gene displayed a statistically significant difference in the risk of DN in T2D patients.Citation16 In addition, they indicated a potential gene-gene interaction between ENPP1 and the growth hormone secretagogue receptor (GHSR) gene using both the generalized multifactor dimensionality reduction method and logistic regression models.Citation16 On the other hand, hypertension is a chronic disorder in which the blood pressure is elevated,Citation18 and hypertension is also the cause of ESRD.Citation1,Citation2 In a population of African Americans, Kao et al showed that MYH9 rs16996677 was highly significantly associated with non-diabetic ESRD and hypertension-associated ESRD.Citation11 Furthermore, another study replicated these results by finding significant associations with MYH9 rs16996677 in non-diabetic ESRD and hypertension-associated ESRD cases among African Americans.Citation13
The MYH9 gene is approximately 110 kb with 41 exons in a region of chromosome 15, and the MYH9 gene product, myosin-IIA, is responsible for moving actin filaments in cells.Citation19,Citation20 Mutations in MYH9 are associated with several Mendelian conditions and autosomal dominant disorders, such as Epstein syndrome, Fechtner syndrome, May-Hegglin anomaly, Sebastian syndrome, and an autosomal dominant form of deafness.Citation21 Kopp et al conducted an admixture-mapping linkage-disequilibrium genome scan and detected an association with MYH9 in African Americans with idiopathic and human immunodeficiency virus-associated focal segmental glomerulosclerosis.Citation12 They also revealed that the MYH9 was significantly associated with non-diabetic forms of ESRD (predominantly ESRD from hypertensive nephrosclerosis), but not with T2D-associated ESRD.Citation12 Kao et al replicated these findings by identifying a highly significant association between MYH9 and several non-diabetic etiologies of ESRD among African Americans but not diabetic ESRD.Citation11 Another study in African Americans further reported that significant associations were detected between MYH9 variants and non-diabetic patients with ESRD (including chronic glomerulonephritis-associated ESRD and hypertension-associated ESRD).Citation13 In a population of African Americans, Freedman et al also demonstrated that MYH9 was associated with T2D-associated ESRD by comparing T2D-ESRD cases with non-diabetic non-nephropathy controls.Citation14 As mentioned previously, Freedman et al did not find an association between MYH9 rs16996677 and kidney failure among African Americans by contrasting the T2D-associated ESRD patients with T2D non-nephropathy controls.Citation14 However, they identified three SNPs, including rs4821480, rs2032487 and rs4821481, that were associated with kidney failure in T2D in the same African American study.Citation14
Moreover, we observed that allele frequencies for the MYH9 rs16996677 SNP in this Taiwanese study were in accordance with HapMap database entriesCitation17 in the populations of European ancestry (that is, CEU) and Asian ancestry (that is, CHB and JPT). However, we did not find a correlation in allele frequency between our study population and HapMap database entriesCitation17 in the populations of African ancestry (that is, ASW and YRI). In addition, MYH9 rs16996677 was not polymorphic among CHB and JPT. This rs16996677 SNP also had only one Taiwanese participant with the heterozygous genotype and none with the rare homozygous genotype.
One limitation of this study is that the contributions of other markers in the MYH9 gene should be further examined in future work. As discussed previously, the selected MYH9 rs16996677 variant was one of the most mentioned SNPs in previous reports.Citation11,Citation13,Citation14 In the current pilot study, we assumed that an SNP might be a good candidate to investigate the genetic role of the implicated gene if the SNP has been investigated in several studies.Citation16 Second, we focused on single locus analyses without considering epistatic models in this study. It is the essential role of gene-gene interactions to define a trait implicating complex disease-related mechanisms, particularly when each involved variant only has a minor marginal effect.Citation22–Citation24 Several statistical methods such as logistic regression and the generalized multifactor dimensionality reduction method have been applied to SNP association studies for detecting gene-gene interactions associated with a number of complex diseases.Citation25–Citation28 In addition, future research using pattern recognition approachesCitation29–Citation31 is needed in order to model associations between gene variants and ESRD. Third, these findings may not be suitably generalized to other populations.Citation16,Citation27 Large ethnically-matched studies would be necessary to know if such an association exists in non-Taiwanese subjects. In future work, we will attempt to recruit more DN and T2D patients as a replication group in a large chronic renal disease and ESRD project for facilitating further analyses.
In conclusion, our study has tested the association between a common SNP rs16996677 in the MYH9 gene and DN in Taiwanese T2D subjects based on single-locus analyses. Our findings did not support the hypothesis that the MYH9 rs16996677 SNP contributes to the risk of DN in T2D. Independent replications in large sample sizes are needed to confirm the role of the polymorphisms found in this study for DN in T2D.
Acknowledgments
The authors extend their sincere thanks to Vita Genomics Inc. for funding this research. The authors would also like to thank the anonymous reviewers for their constructive comments, which improved the context and presentation of this paper.
Disclosures
The authors have no conflicts of interest to disclose.
References
- GrassmannAGiobergeSMoellerSBrownGESRD patients in 2004: Global overview of patient numbers, treatment modalities and associated trendsNephrol Dial Transplant200520122587259316204281
- MeguidElNahasABelloAKChronic kidney disease: The global challengeLancet2005365945633134015664230
- RemuzziGSchieppatiARuggenentiPClinical practice. Nephropathy in patients with type 2 diabetesN Engl J Med2002346151145115111948275
- StumvollMGoldsteinBJvan HaeftenTWType 2 diabetes: Principles of pathogenesis and therapyLancet200536594671333134615823385
- HattersleyATPearsonERMinireview: Pharmacogenetics and beyond: The interaction of therapeutic response, beta-cell physiology, and genetics in diabetesEndocrinology200614762657266316556760
- FreedmanBIBostromMDaeihaghPBowdenDWGenetic factors in diabetic nephropathyClin J Am Soc Nephrol2007261306131617942768
- de BorstMHBenigniARemuzziGPrimer: Strategies for identifying genes involved in renal diseaseNat Clin Pract Nephrol20084526527618364721
- ConwayBRMaxwellAPGenetics of diabetic nephropathy: Are there clues to the understanding of common kidney diseases?Nephron Clin Pract20091124c213c22119546580
- LalwaniAKGoldsteinJAKelleyMJLuxfordWCasteleinCMMhatreANHuman nonsyndromic hereditary deafness DFNA17 is due to a mutation in nonmuscle myosin MYH9Am J Hum Genet20006751121112811023810
- Even-RamSYamadaKMOf mice and men: Relevance of cellular and molecular characterizations of myosin IIA to MYH9-related human diseaseCell Adh Migr20071315215519262128
- KaoWHKlagMJMeoniLAMYH9 is associated with non-diabetic end-stage renal disease in African AmericansNat Genet200840101185119218794854
- KoppJBSmithMWNelsonGWMYH9 is a major-effect risk gene for focal segmental glomerulosclerosisNat Genet2008401011751178418794856
- FreedmanBIHicksPJBostromMAPolymorphisms in the non-muscle myosin heavy chain 9 gene (MYH9) are strongly associated with end-stage renal disease historically attributed to hypertension in African AmericansKidney Int200975773674519177153
- FreedmanBIHicksPJBostromMANon-muscle myosin heavy chain 9 gene MYH9 associations in African Americans with clinically diagnosed type 2 diabetes mellitus-associated ESRDNephrol Dial Transplant200924113366337119567477
- FreedmanBIKoppJBWinklerCAPolymorphisms in the nonmuscle myosin heavy chain 9 gene (MYH9) are associated with albuminuria in hypertensive African Americans: The HyperGEN studyAm J Nephrol200929662663219153477
- WuLSHsiehCHPeiDHungYJKuoSWLinEAssociation and interaction analyses of genetic variants in ADIPOQ, ENPP1, GHSR, PPAR{gamma} and TCF7L2 genes for diabetic nephropathy in a Taiwanese population with type 2 diabetesNephrol Dial Transplant200924113360336619506043
- The International HapMap ConsortiumThe International HapMap ProjectNature2003426696878979414685227
- CarreteroOAOparilSEssential hypertension. Part I: definition and etiologyCirculation2000101332933510645931
- D’ApolitoMGuarnieriVBoncristianoMZelanteLSavoiaACloning of the murine non-muscle myosin heavy chain IIA gene ortholog of human MYH9 responsible for May-Hegglin, Sebastian, Fechtner, and Epstein syndromesGene2002286221522211943476
- MariniMBruschiMPecciANon-muscle myosin heavy chain IIA and IIB interact and co-localize in living cells: Relevance for MYH9-related diseaseInt J Mol Med200617572973616596254
- SinghNNainaniNAroraPVenutoRCCKD in MYH9-related disordersAm J Kidney Dis200954473274019726116
- LinEHwangYLiangKHChenEYPattern-recognition techniques with haplotype analysis in pharmacogenomicsPharmacogenomics200781758317187511
- LinEHwangYChenEYGene-gene and gene-environment interactions in interferon therapy for chronic hepatitis CPharmacogenomics20078101327133517979507
- LinEHsuSYA Bayesian approach to gene-gene and gene-environment interactions in chronic fatigue syndromePharmacogenomics2009101354219102713
- LouXYChenGBYanLA generalized combinatorial approach for detecting gene-by-gene and gene-by-environment interactions with application to nicotine dependenceAm J Hum Genet20078061125113717503330
- LinEHongCJHwangJPGene-gene interactions of the brain-derived neurotrophic-factor and neurotrophic tyrosine kinase receptor 2 genes in geriatric depressionRejuvenation Res200912638739320014955
- LinEPeiDHuangYJHsiehCHWuLSGene-gene interactions among genetic variants from obesity candidate genes for nonobese and obese populations in type 2 diabetesGenet Test Mol Biomarkers200913448549319594364
- LinEChenPSChangHHInteraction of serotonin-related genes affects short-term antidepressant response in major depressive disorderProg Neuropsychopharmacol Biol Psychiatry20093371167117219560507
- LinEHwangYWangSCGuZJChenEYAn artificial neural network approach to the drug efficacy of interferon treatmentsPharmacogenomics2006771017102417054412
- LinEHuangLCIdentification of significant genes in genomics using Bayesian variable selection methodsAdvances and Applications in Bioinformatics and Chemistry20081131821918603
- LinEHwangYA support vector machine approach to assess drug efficacy of interferon and ribavirin combination therapyMol Diagn Ther200812421922318652518