Abstract
Aim: To assess whether expression of circular RNAs (circRNAs) in peripheral blood mononuclear cells can serve as a biomarker for diagnosis and/or therapeutic response in people living with schizophrenia (SZ). Materials & methods: Differentially expressed circRNAs were screened via microarray in nine individuals living with SZ and nine healthy controls, then quantified using real-time quantitative reverse transcription PCR in SZ (n = 102) and healthy control (n = 103) groups. CircRNAs were re-assessed twice in 30 randomly selected individuals living with SZ after 4- and 8-week antipsychotic treatments. Results: Five circRNAs were differentially expressed between groups. Only hsa_circRNA_104597, which was downregulated in the SZ group, was significantly upregulated after 8-week treatment. Conclusion: Dysregulation of hsa_circRNA_104597 may serve as a novel potential diagnostic and therapeutic biomarker for SZ.
Schizophrenia (SZ) is a serious neuropsychiatric disorder with high recurrence and disability rates. McGrath et al. reported that the lifetime prevalence, annual average morbidity and lifetime morbidity risk were 0.46, 0.015 and 0.72%, respectively [Citation1]. Under less stringent diagnostic criteria, these rates grow as much as twofold [Citation2]. An inability to maintain employment and a lower life expectancy with SZ create a severe economic and social burden. Current evidence indicates that SZ is caused by an interaction between environmental and genetic factors, although its pathogenesis remains unclear. Furthermore, a lack of reliable biomarkers hampers the early diagnosis and effective treatment of people living with SZ [Citation3].
Circular RNA (circRNA) is a eukaryotic RNA species first found in plants in the 1970s [Citation4]. Its native form is circular [Citation5,Citation6], which makes it highly stable and resistant to RNase R degradation [Citation7]. CircRNAs function as competing endogenous RNA, and as such may be involved in disease progression [Citation8–10]. The hypothesis of competing endogenous RNA-gained substantial attention as a potential unifying function for mRNA, long noncoding RNA, pseudogene transcripts and circRNA. Given the stability of circRNA, circRNAs may surpass linear RNAs as miRNA sponges [Citation11,Citation12]. Furthermore, circRNAs may potentially serve as minimally invasive biomarkers for disorders of CNS, since they can freely cross the blood–brain barrier [Citation13]. Recent studies suggest that the circRNA Cdr1as is massively bound by the miRNAs such as miR-7 and miR-671 in human and mouse brains [Citation14], and the Cdr1as genome knockout mice displayed impaired sensorimotor gating-a deficit in the ability to filter out unnecessary information, which is associated with human neuropsychiatric disorders [Citation15]. Considering a strong deficit in prepulse inhibition of individuals diagnosed with SZ, we supposed that some circRNAs might participate in the pathogenesis and development of SZ.
Despite the growing evidence that circRNAs might participate in the mechanisms underlying SZ pathophysiology, no data have been published regarding the direct relationship between circRNA and SZ. Furthermore, Cdr1as is highly abundant only in the mammalian brain and expressed at low levels or absent in other tissues and organs [Citation14]. That makes it hard to serve as a minimally invasive biomarker. To assess whether expression of circRNAs in peripheral blood mononuclear cells (PBMCs) can serve as a minimally invasive biomarker for diagnosis and/or therapeutic response in people living with SZ, we analyzed circRNA expression profiles in PBMCs from individuals living with SZ and healthy controls (HCs). We also analyzed changes in expression related to the treatment of individuals living with SZ with antipsychotic medications.
Materials & methods
Subjects
In order to keep the same diagnostic criteria with our previous study for the following study regarding the interaction of circRNAs and miRNAs [Citation16,Citation17]. Individuals diagnosed with SZ (outpatient or inpatient) according to the criteria of the Diagnostic and Statistical Manual of Mental Disorders, 4th Edition (DSM-IV) [Citation18] were enrolled from No.904 Hospital of the Chinese People’s Liberation Army (Changzhou, China) between August 2014 and October 2015. Inclusion criteria were: first visitors and no history of clinical treatment for SZ or no antipsychotic treatment for at least 3 months before blood sampling; no other disease, recreational drug use or history of blood transfusion 1 month before study enrollment and age 16–65 years. Exclusion criteria included: serious organic disease, affective disorder or other psychiatric disorders; personality disorder from sequential medicine; alcohol abuse or other substance abuse; modified electroconvulsive therapy within 6 months or long-term use of antipsychotic drugs; need for long-term combined use of mood-altering or CNS-acting drugs. In total, 102 individuals living with SZ (44 men and 58 women), with a mean age of 33.41 ± 15.3 years were enrolled.
HCs were recruited from the hospital staff. Inclusion criteria were: ethnic Han; age 16–65 years; no diagnosis of SZ or other mental disease following a structured clinical interview per the DSM-IV, nonpatient edition (SCID-I/NP). Controls were also excluded in cases of a family history of major psychiatric disorders (SZ, bipolar disorder or major depressive disorder) within at least two generations, serious nervous system disease, other physical disease or a history of severe traumatic event or blood transfusion within a month before blood sampling. In total, 103 HCs were selected (49 men, 54 women; mean age 35.62 ± 13.9 years [range, 18–62]). The medical ethics board of No. 904 Hospital approved the study and written informed consent was obtained from all the individuals.
RNA extraction & circRNA microarray expression profiling
Venous blood (5 ml) was collected in EDTA anticoagulant tubes and centrifuged (3000 × g for 10 min) within 3 h. PBMCs were isolated from the blood through density gradient centrifugation, then transferred into 2-ml fresh RNase/DNase-free tubes and stored at -80°C until use. Total RNA was isolated from the PBMCs using an RNeasy Mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol, and quantified on a NanoDrop ND-1000 (Thermo Scientific, ME, USA). RNA integrity was evaluated after agarose gel electrophoresis.
Referring to the sample size for microarray matrix of previous studies [Citation19,Citation20] and using sex- and age-stratified sampling, blood samples from nine of the 102 individuals living with SZ (men, ages 23, 35, 47 and 62 years; women, 19, 28, 40, 49 and 64 years) and nine age- and sex-matched HCs (men, ages 22, 35, 46 and 61 years; women, 19, 28, 40, 48 and 64 years) out of the 103 healthy controls were collected. After quality inspection, qualified samples were labeled with Cy3-CTP (Cy3) and hybridized onto an Arraystar Human circRNA Array (8x15K, Arraystar, MD, USA). The elution of RNAs was performed according to the manufacturer’s protocols. Hybrid images were obtained on an Agilent G2505C Scanner (Agilent Technologies, CA, USA).
Agilent Feature Extraction software (version 11.0.1.1) was used to analyze acquired array images. Quantile normalization, ComBat adjustment and subsequent data processing were performed using the R software package. Volcano Plot filtering was used to identify circRNAs with statistically significant between group differences in expression, and hierarchical clustering to distinguish the circRNA expression pattern among samples.
Reverse transcription PCR
Nine circRNAs with statistically significant differences between groups were isolated and selected for further validation using real-time quantitative reverse transcription-PCR (qRT-PCR). Blood samples from the total group populations were used to validate the findings from circRNA profiling. Complementary DNA (cDNA) was transcribed using a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Inc., USA), and circRNA fluorescence probe was synthesized by Invitrogen (Thermo Fisher Scientific, USA). A TaqMan reagent kit (TaqMan General Mixed Reagent kit II, ABI, USA) was used to perform real-time PCR. The cycle threshold (Ct) was obtained and relative expression levels of the tested circRNAs were calculated based on the following formula using β-actin as a normalization control: 2-△Ct (△Ct = CtcircRNA – Ctβ-actin). Greater △Ct values represent lower actual expression levels.
Medical intervention
Thirty individuals living with SZ were randomly selected from the total SZ enrollment. These individuals received olanzapine monotherapy (5 mg daily, gradually increased to 10–20 mg daily). If extrapyramidal symptoms occurred, the individual received benzhexol hydrochloride orally. CircRNA expression in these 30 individuals was re-assessed twice as described above at 4 and 8 weeks after the initiation of olanzapine.
Statistical analysis
Sociodemographic variables were compared between the SZ and HC groups by t-test for continuous variables and by χ2 test for categorical variables. Volcano Plot filtering was performed using a threshold of fold change ≥1.5 and p-value < 0.05. The Mann–Whitney U-test was used to compare expression between groups. Changes in expression between the 0-, 4- and 8-week olanzapine administration time points were analyzed using repeated measures analysis of variance (ANOVA) followed by Bonferroni’s post hoc test for multiple comparisons if significant changes were seen. The receiver operating characteristics (ROC) curve was established to evaluate potential diagnostic value.
CircRNA expression is reported as means ± standard error of the mean; all data were processed by DataAssist v3.0 and SPSS version 20.0 (IBM, IL, USA). All statistical tests were two-tailed, and p < 0.05 was considered statistically significant.
Results
Demographic characteristics
As shown in , no statistically significant differences in age and sex (p = 0.52 and 0.28, respectively) were found between the SZ and HC groups.
Table 1. Demographic characteristics of individuals living with schizophrenia and healthy controls.
CircRNA microarray screening
Using the screening standard defined in the Methods section, nine circRNAs were differentially expressed ( & ). Of these, six were upregulated and three were downregulated.
The volcano plot was constructed using fold change values and p-values. The vertical lines correspond to 1.5-fold up- and down-regulation between individuals living with SZ and HC samples, and the horizontal line represents a p-value of 0.05. The red points in the plot represent the statistically significant, differentially expressed circular RNAs.
HC: Healthy control SZ: Schizophrenia.
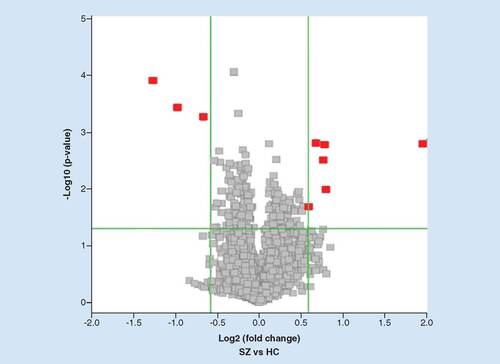
Table 2. Circular RNAs with altered expressions screened by microarray.
Cluster analysis of differentially expressed circRNAs
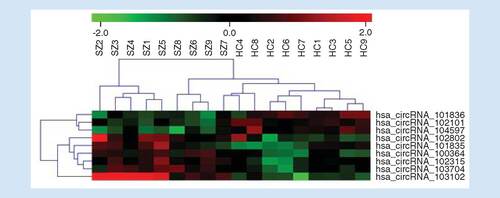
Hierarchical clustering under the Euclidean metric and complete linkage rule was applied to generate a heat map of the normalized microarray expression data of the nine circRNAs demonstrating differential expression between groups ().
Rows represent circular RNA species, and columns represent individual samples. Color gradations are used to represent the expression levels of circRNAs (red for high-expression and green for low-expression). The numbers with SZ denote individuals living with schizophrenia while HC denote healthy controls.
HC: Healthy control; SZ: Schizophrenia.
Validation of circRNA expression by qRT-PCR
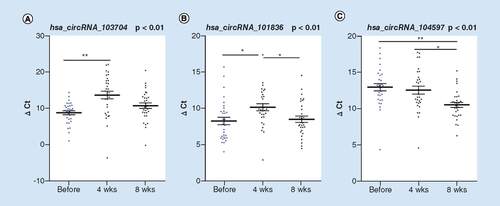
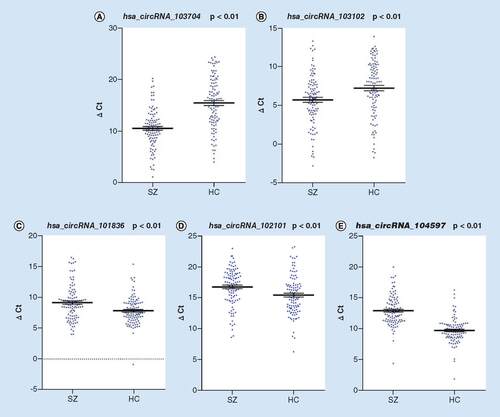
Using β-actin as the internal reference, the above nine circRNAs were validated against the full 205 sample group. A Mann–Whitney U-test on two independent samples showed there was no significant difference in △Ct values of hsa_circRNA_102802 and hsa_circRNA_100364 between the two groups (Z = -1.053 and -0.732; p = 0.2931 and 0.4647). The expression trends of hsa_circRNA_101835 and hsa_circRNA_102315 were reversed relative to those of the microarray and thus excluded from further analysis. Of the five remaining circRNAs (), the △Ct values of hsa_circRNA_101836, hsa_circRNA_102101, and hsa_circRNA_104597 in the SZ group were significantly higher (expression was significantly downregulated) than those in the HC group (Z = -3.457, -3.405 and -9.520; p = 0.0005, 0.0007 and p < 0.0001, respectively). Conversely, the expression levels of hsa_circRNA_103704 and hsa_circRNA_103102 in the SZ group were significantly upregulated compared with the HC group (Z = -6.961 and -3.334; p < 0.0001 and 0.0009).
The figure shows △Ct of the five circRNAs that were significantly different between the individuals living with SZ and HC groups. Lines and error bars represent mean and standard error of the mean.
circRNA: Circular RNA; HC: Healthy control; SZ: Schizophrenia.
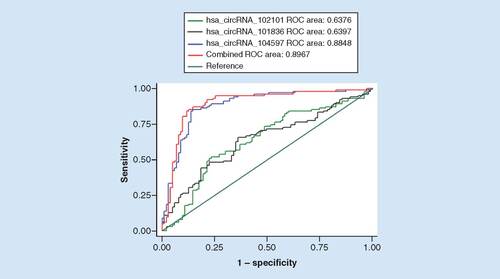
In analyzing the ROC curve (), the area under the curve values of hsa_circRNA_102101, hsa_circRNA_104597 and hsa_circRNA_101836 were 0.6376 (95% CI: 0.561–0.715), 0.8848 (95% CI: 0.837–0.933) and 0.6397 (95% CI: 0.564–0.716), respectively. When all three circRNAs were used to create a combined ROC curve, the area under the curve was 0.8967 (95% CI: 0.861–0.953), with 87.25% sensitivity and 85.44% specificity.
Comparison of circRNA expression across sex & age
According to WHO age segmentation guidelines, the 102 individuals living with SZ were divided into two subgroups, youth (≤44 years, n = 73) and middle-aged (>44 years, n = 29). A Mann–Whitney U-test on two independent samples revealed that there were no differences in the expression levels of the five circRNAs between the two age subgroups (Z = -1.673 to -0.056; p = 0.094 to 0.956), or sex (Z = -1.142 to -0.108; p = 0.254 to 0.914).
Comparison of circRNA expression after 0, 4 & 8 weeks of olanzapine monotherapy
Repeated measures one-way ANOVA revealed significant differences between the 0- and 8-week time points in hsa_circRNA_103704 (F(2, 58) = 9.395; p = 0.0003), hsa_circRNA_101836 (F(2, 58) = 5.086; p = 0.0092) and hsa_circRNA_104597 (F(2, 58) = 8.011; p = 0.0008). Bonferroni’s post hoc test for multiple comparisons identified significant downregulation between the 0- and 4-week time points in expression levels of hsa_circRNA_103704 (p = 0.0005), and hsa_circRNA_101836 (p = 0.0446), but not hsa_circRNA_104597 (p = 1.0000); between the 4- and 8-week time points, significant upregulation of expression was identified for hsa_circRNA_101836 (p = 0.0333) and hsa_circRNA_104597 (p = 0.0141), but not hsa_circRNA_103704 (p = 0.0751). Between the 0- and 8-week time points, only hsa_circRNA_104597 expression was significantly different (p = 0.0002) ().
Discussion
In the present study, we systematically screened circRNAs that were differentially expressed between individuals living with SZ and HCs. Our results indicate that several known circRNAs may correlate with the expression of SZ. The array data will be available when we finish the following study. Evidence suggests that circRNA may play an important role in the development of SZ, with Cdr1as in particular having support [Citation15]. However, the low levels or absent in peripheral tissues reduces Cdr1as’ capability to serve as a minimally invasive biomarker [Citation14]. Therefore, we hypothesized that certain circRNAs, which is abundant in PBMCs, also have a potential role in the development of SZ. We found nine mature circRNAs to be expressed differently between groups, of which six were upregulated and three downregulated. After validation against the full 205 participants’ samples, three of the circRNAs (hsa_circRNA_101836, hsa_circRNA_102101 and hsa_circRNA_104597) were downregulated and two circRNAs (hsa_circRNA_103704 and hsa_circRNA_103102) were upregulated in the SZ group relative to the HC group.
Research on blood biomarkers of SZ has tested CRP [Citation21], cytokines [Citation22,Citation23], hormones (e.g., cortisol, insulin, leptin, prolactin and growth hormone) [Citation24], miRNAs [Citation25,Citation26] and long noncoding RNAs [Citation27], but all demonstrated low sensitivity and/or specificity. Our ROC curve analysis showed three of the circRNAs were diagnostically valuable, with a sensitivity of 87.25% and specificity of 85.44%. Notably, hsa_circRNA_104597 alone had a sensitivity of 84.31% and specificity of 86.41%. Thus, our results suggest the three circRNAs, particularly hsa_circRNA_104597, have potential as diagnostic biomarkers of SZ.
SZ demonstrates sex and age biases, including higher morbidity in women and the elderly. A previous study indicated that dynamic changes in circRNA expression were both sex- and age-biased in rhesus macaque brains [Citation28]. In addition, small noncoding RNA expression in SZ is regulated sex-specifically [Citation29,Citation30]. Our data indicated no sex- or age-based dysregulation of the five circRNAs in PBMCs from patients living with SZ patients, but further studies with larger sample sizes are needed to confirm this.
The ‘perfect’ biomarker should have prognostic value as well. However, most previous studies on ncRNAs evaluated expression between SZ and HC groups, but not between pretreated and treated SZ groups. We found the expression level of hsa_circRNA_104597 to be downregulated before treatment, then upregulated after treatment for 8 weeks. Although the underlying mechanisms remain unknown, our results suggest that hsa_circRNA_104597 might be a potential therapeutic biomarker for SZ.
MiRNA prediction software developed by Arraystar based on TargetScan [Citation31] and miRanda [Citation32] was used to predict the interaction between the circRNAs and miRNAs. Hsa_circRNA_104597 contained five binding sites for hsa-miR-659-3p, hsa-miR-548d-5p, hsa-miR-651-3p, hsa-miR-548c-5p and hsa-miR-548a-5p. Studies indicate the five miRNAs were associated with the neural system, cardiovascular system and cancer [Citation33–36]. Furthermore, genetic studies indicate abnormalities in the 8p, 13q and 22q chromosome arms are implicated in the pathogenesis of SZ [Citation37]; hsa_circRNA_104597 is found on chromosome 8.
Conclusion
CircRNAs in PBMCs represent a promising, minimally invasive source for the development of diagnostic and therapeutic tools. We identified five circRNAs that might serve as such biomarkers for SZ. Our results will be beneficial for research on the pathogenesis of SZ and suggest hsa_circRNA_104597 specifically could serve as a blood-based biomarker.
Limitations
There are some limitations to this study. First of all, the array data were collected from a small sample of only nine individuals living with SZ and nine healthy controls, and require further validation. Second, there is a potential for selection bias in the 30-individual subgroup that is difficult to avoid. Based on ethical considerations, we did not include a placebo group, so a time-dependent alteration in circRNA expression cannot be eliminated. Next, the healthy controls were hospital staff members and may not represent the general population. Finally, the short therapeutic treatment time may not truly represent circRNA expression changes before and after long-term therapy. A study with a longer follow-up time is needed to clarify the influence of medical treatment on the changes in circRNA expression.
Methodological issues in the assessment of circular RNAs with altered expression
Differentially expressed circular RNAs (circRNAs) were screened between individuals living with schizophrenia (SZ) and healthy controls using the Arraystar Human CircRNA Array.
Reverse transcription-PCR was used to amplify target genes and compare the differences in gene expression.
Diagnostic value of circRNAs for SZ
Nine circRNAs with altered expression were identified in peripheral blood mononuclear cells from individuals living with SZ using the Arraystar Human CircRNA Array.
Five circRNAs were confirmed to be significantly different between individuals living with SZ and healthy controls after validation by reverse transcription-PCR.
No significant difference in the expression levels of the five circRNAs were observed between different sex and age.
Three circRNAs demonstrated a diagnostic value for SZ using receiver operating characteristics curve analysis. In particular, hsa_circRNA_104597 had a strong diagnostic value, with an area under the curve value of 0.8848 (95% CI: 0.837–0.933).
Therapeutic value of circRNAs for SZ
Only the expression levels of hsa_circRNA_104597 in peripheral blood mononuclear cells from individuals living with SZ showed significant changes after 8-week antipsychotic treatment with olanzapine monotherapy.
Author contributions
G-F Yao performed main analyses, wrote the draft and made interpretation and critical revision in the final version of the manuscript. W Niu collected and organized the data, and participated in the critical discussion of the study design. X-L Zhu and M He performed main laboratory work and interpretation of data regarding to intellectual content. L-M Kong and S-D Chen collected and organized the information data. L-Y Zhang and Z-H Cheng conceived and designed the study, participated in the diagnoses and enrollment of the patients, and were also responsible for the critical revision of the manuscript.
Ethical conduct of research
The medical ethics board of No.904 Hospital approved the study and written informed consent was obtained from all the individuals. The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Data sharing statement
The authors certify that this manuscript reports original clinical trial data (Clinical Trial registration no.: ChiCTR-OOC-16007994). Individual participant data that underlie the results reported in this article, after de-identification (text, table, figures and appendices), along with the study protocol, will be available beginning 3 months and ending 24 months following article publication. This information will be available to researchers who provide a methodologically sound proposal, to the achieve aims in the approved proposal. Proposals should be directed to [email protected]. To gain access, data requestors will need to sign a data access agreement. Data are available for 24 months at a third party website (www.chictr.org.cn).
Acknowledgements
We wish to convey sincere thanks to all the subjects who participated in the study. We also wish to thank Gopath Global LLC., Chicago in USA for their professional laboratory services and No. 904 Hospital clinical laboratory of PLA for professional laboratory assistance.
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
- McGrath J , SahaS, ChantDet al. Schizophrenia: a concise overview of incidence, prevalence, and mortality. Epidemiol. Rev.30, 67–76 (2008).
- van Os J , KapurS. Schizophrenia. Lancet374(9690), 635–645 (2009).
- Insel TR . Disruptive insights in psychiatry: transforming a clinical discipline. J. Clin. Invest.119(4), 700–705 (2009).
- Sanger HL , KlotzG, RiesnerD. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl Acad. Sci. USA73(11), 3852–3856 (1976).
- Capel B , SwainA, NicolisSet al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell73(5), 1019–1030 (1993).
- Cocquerelle C , DaubersiesP, MajerusMAet al. Splicing with inverted order of exons occurs proximal to large introns. EMBO J.11(3), 1095–1098 (1992).
- Suzuki H , KameyamaT, OheKet al. Nested introns in an intron: evidence of multi-step splicing in a large intron of the human dystrophin pre-mRNA. FEBS Lett.587(6), 555–561 (2013).
- Salzman J , GawadC, WangPLet al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE7(2), e30733 (2012).
- Hansen TB , JensenTI, ClausenBHet al. Natural RNA circles function as efficient microRNA sponges. Nature495(7441), 384–388 (2013).
- Salmena L , PolisenoL, TayYet al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146(3), 353–358 (2011).
- Memczak S , JensM, ElefsiniotiAet al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature495(7441), 333–338 (2013).
- Zhuang CL , FuX, LiuLet al. Synthetic miRNA sponges driven by mutant hTERT promoter selectively inhibit the progression of bladder cancer. Tumour Biol.36(7), 5157–5163 (2015).
- Lu D , XuAD. Mini review: circular RNAs as potential clinical biomarkers for disorders in the central nervous system. Front. Genet.7, 53 (2016).
- Hansen TB , WiklundED, BramsenJBet al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO. J.30(21), 4414–4422 (2011).
- Piwecka M , GlažarP, Hernandez-MirandaLRet al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science357(6357) (2017).
- Sun XY , ZhangJ, NiuWet al. A preliminary analysis of microRNA as potential clinical biomarker for schizophrenia. Am. J. Med. Genet. B Neuropsychiatr. Genet.168B(3), 170–178 (2015).
- Song HT , SunXY, ZhangLet al. A preliminary analysis of association between the down-regulation of microRNA-181b expression and symptomatology improvement in schizophrenia patients before and after antipsychotic treatment. J. Psychiatr. Res.54, 134–140 (2014).
- American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders, 4th ed. (DSM-IV). AmericanPsychiatric Association Publishing. (1994).
- Cui X , NiuW, KongLet al. hsa_circRNA_103636: potential novel diagnostic and therapeutic biomarker in major depressive disorder. Biomark. Med.10(9), 943–952 (2016).
- Zheng F , YuX, HuangJ, DaiY. Circular RNA expression profiles of peripheral blood mononuclear cells in rheumatoid arthritis patients, based on microarray chip technology. Mol. Med. Rep.16(6), 8029–8036 (2017).
- Wang Z , LiP, ChiDet al. Association between C-reactive protein and risk of schizophrenia: an updated meta-analysis. Oncotarget8(43), 75445–75454 (2017).
- Hartwig FP , BorgesMC, HortaBLet al. Inflammatory biomarkers and risk of schizophrenia: a 2-sample Mendelian Randomization Study. JAMA Psychiatry74(12), 1226–1233 (2017).
- Schwarz E , GuestPC, RahmouneHet al. Identification of a blood-based biological signature in subjects with psychiatric disorders prior to clinical manifestation. World J. Biol. Psychiatry13(8), 627–632 (2012).
- Harris LW , PietschS, ChengTMet al. Comparison of peripheral and central schizophrenia biomarker profiles. PLoS ONE7(10), e46368 (2012).
- Ma J , ShangS, WangJet al. Identification of miR-22-3p, miR-92a-3p, and miR-137 in peripheral blood as biomarker for schizophrenia. Psychiatry Res.265, 70–76 (2018).
- Liu S , ZhangF, WangXet al. Diagnostic value of blood-derived microRNAs for schizophrenia: results of a meta-analysis and validation. Sci. Rep.7(1), 15328 (2017).
- Chen S , SunX, NiuWet al. Aberrant expression of long non-coding RNAs in schizophrenia patients. Med. Sci. Monit.22, 3340–3351 (2016).
- Xu K , ChenD, WangZet al. Annotation and functional clustering of circRNA expression in rhesus macaque brain during aging. Cell Discov.4, 48 (2018).
- Ragan C , PatelK, EdsonJet al. Small non-coding RNA expression from anterior cingulate cortex in schizophrenia shows sex specific regulation. Schizophr. Res.183, 82–87 (2017).
- Safari MR , KomakiA, Arsang-JangSet al. Expression pattern of long non-coding RNAs in schizophrenic patients. Cell Mol. Neurobiol.doi: https://doi.org/10.1007/s10571-018-0640-3 (2018) ( Epub ahead of print).
- Enright AJ , JohnB, GaulUet al. MicroRNA targets in Drosophila. Genome Biol.5(1), R1. (2003)
- Pasquinelli AE . MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet.13(4), 271–282 (2012).
- Li Y , XieJ, XuXet al. MicroRNA-548 down-regulates host antiviral response via direct targeting of IFN-λ1. Protein Cell4(2), 130–141 (2013).
- Gupta MK , HalleyC, DuanZHet al. miRNA-548c: a specific signature in circulating PBMCs from dilated cardiomyopathy patients. J. Mol. Cell Cardiol.62, 131–141 (2013).
- Rademakers R , EriksenJL, BakerMet al. Common variation in the miR-659 binding-site of GRN is a major risk factor for TDP43-positive frontotemporal dementia. Hum. Mol. Genet.17(23), 3631–3642 (2008).
- Pu X , RothJA, HildebrandtMAet al. MicroRNA-related genetic variants associated with clinical outcomes in early-stage non-small cell lung cancer patients. Cancer Res.73(6), 1867–1875 (2013).
- Badner JA , GershonES. Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol. Psychiatry7(4), 405–411 (2002).