Abstract
It is important to clarify the significance of long noncoding RNA MIR31 host gene (lncRNA MIR31HG) in predicting the prognosis for malignant tumors through meta-analysis. Electronic databases were systemically searched, from inception until 2 January 2019, to identify related articles. Meanwhile, the hazard ratios (odds ratios) and 95% CIs were computed for exploring the association between the expression of lncRNA MIR31HG and the survival (pathological variables). Eleven studies with 1041 cases were enrolled into the current meta-analysis. Low expression of lncRNA MIR31HG showed correlation with the dismal overall survival, disease-free survival, high tumor stage and lymph node metastasis among patients with digestive system cancers. Low expression of lncRNA MIR31HG may serve as a potential novel factor to indicate the dismal prognosis and metastasis in patients with digestive system cancers.
Keywords:
According to reports in recent years, there are approximately 1.7 million newly diagnosed cancer patients in the USA in 2017, and the cancer-associated deaths are 606,880 [Citation1]. However, the 5-year survival for a majority of malignancies is still quite unsatisfactory; meanwhile, numerous scientists have made tremendous efforts to search for the novel biomarkers for determining or predicting the prognosis for cancer.
LncRNA, in which the meaningful open-reading frame (ORF) is lacking, has been recognized to be the transcribed RNA molecule that is greater than 200 nucleotides in length. It plays numerous vital roles in disease, for instance, regulation at post-transcriptional, transcriptional as well as epigenetic levels [Citation2,Citation3]. Additionally, it is suggested that the aberrant expression of lncRNA is correlated with a variety of cancer types [Citation4–Citation7]. For example, some lncRNAs play vital roles in cancer cell metastasis, invasion as well as proliferation, which suggests that lncRNA can be used to be an effective marker to predict the prognosis for cancer [Citation8–Citation10].
Long noncoding RNA MIR31 host gene (lncRNA MIR31HG) is one of these lncRNAs, which is localized in chromosome 9p21.3 with the length of 2166 bp. In addition, recent studies reveal that the transcription of lncRNA MIR31HG is regulated by the methylation in the promoter region, and lncRNA MIR31HG has contributed an important role in numerous biological processes, including migration, proliferation and apoptosis [Citation11–Citation13]. LncRNA MIR31HG may affect the metastasis and prognosis for cancer; however, most current research is restricted due to the small sample size as well as the discrete outcomes. Therefore, this study carried out a meta-analysis to examine the significance of lncRNA MIR31HG in predicting the prognosis for cancer patients.
Materials & methods
Study retrieval
With reference to the classical guidelines of meta-analysis [Citation14,Citation15], electronic databases (including Web of Science database, PubMed and OVID) were independently systemically searched from inception to 2 January 2019 by two reviewers, so as to identify the relevant studies that examined the role of lncRNA MIR31HG in predicting the prognosis for cancer patient survival. At the same time, the text words as well as the Mesh strategies were adjusted according to the retrieved databases in this study. The following terms were used in this study, including (‘long non-coding RNA MIR31 host gene’ or ‘LncRNA MIR31HG’ or ‘LncRNA LOC554202’) and (‘recurrence’ or ‘outcome’ or ‘survival’, ‘cancer’ or ‘neoplasm’ or ‘tumor’ or ‘carcinoma’, ‘prognosis’ or ‘prognostic’). In addition, related references listed in relevant studies were also searched manually, so as not to miss any potentially qualified study.
Study screening
Afterward, each of the enrolled articles was assessed, and the interest data were collected by two authors independently. Studies meeting the following inclusion criteria were enrolled: tumors were verified histologically or pathologically; expression of lncRNA MIR31HG was detected in human tumor tissues; cases were divided based on the expression of lncRNA MIR31HG; there were enough original data to carry out statistical analyses on the pathological parameters or patient survival and the expression of lncRNA MIR31HG.
Besides, studies conforming to the following exclusion criteria were excluded: nonhuman articles or non-English articles; editorials, reviews, expert opinions or letters; analysis of database with no original data; articles that only examined the functions as well as the molecular structure of lncRNA MIR31HG.
Date collection
Data were evaluated and collected from the original studies through two reviewers independently. Any disagreement between them in the literature evaluation was solved through the opinion of a third reviewer. The following data were extracted for meta-analysis, namely, the surname of the first author, year of publication, region, tumor type, sample size, the case number with large tumor size (LTS), poor histological grade (PHG), high tumor stage (HTS), lymph node metastasis (LNM) as well as distant metastasis (DM), reference gene, together with the method for detecting lncRNA MIR31HG, the hazard ratios (HRs) with the corresponding 95% CIs regarding the upregulated expression of lncRNA MIR31HG for the overall survival (OS) as well as disease-free survival (DFS).
Statistical methods
Statistical analysis was performed using the Stata12.0 software. The heterogeneities were detected in the current meta-analysis by Q and I2 tests, and the results revealed that there was an obvious heterogeneity in the current study (I2 ≥50%, and p < 0.1) [Citation16], so the random effect model was selected. On the other hand, Egger’s test as well as Begg’s funnel plot was employed to evaluate the possible publication bias. The aggregated odds ratios (ORs) as well as the HRs were collected from the published studies, and the original data of HRs were collected directly from the original data if they were directly available in the published studies. Additionally, survival data were collected based on the Kaplan–Meier curves, so as to predict the related HRs not directly available from the studies. Then, the log HRs were adopted to summarize the survival outcomes [Citation17]. In addition, the 95% CIs, together with ORs, were used in combination to evaluate the associations between the clinicopathological parameters and the expression of lncRNA MIR31HG.
Results
Characteristics of the enrolled studies
The literature selection process is displayed in . Eleven studies with 1041 cases were recruited into the current meta-analysis according to the exclusion as well as inclusion criteria [Citation18–Citation28]. illustrates the enrolled study characteristics in the current meta-analysis. The table suggested that the sample size of these articles published from 2015 to 2018 in China was 42–185 (average, 94.64). One respective article concentrated on the cervical cancer (CVR) [Citation18], bladder cancer (BC) [Citation20], gastric cancer (GC) [Citation22], lung adenocarcinoma (LADC) [Citation23], esophageal squamous cell carcinoma (ESCC) [Citation24], oral cancer (ORC) [Citation25], laryngeal squamous cell cancer (LSCC) [Citation26] and hepatocellular carcinoma (HCC) [Citation27], while three on colorectal cancer (CRC) [Citation19,Citation21,Citation28]. Moreover, all clinical pathological parameters were pathology dependent, and the lncRNA MIR31HG reference genes were inconsistent among the collected articles, including glyceraldehyde-3-phosphate dehydrogenase (GAPDH) [Citation18–Citation20,Citation22–Citation25,Citation27], β-actin [Citation28] and 18s rRNA [Citation26].
Table 1. The basic information and data of all included studies in the meta-analysis.
Relationship of the expression of lncRNA MIR31HG with OS
Cumulative meta-analysis was performed in the current meta-analysis, so as to evaluate the effect of lncRNA MIR31HG on the OS for cancer patients. It was observed that nine articles with 938 cases reported the association between OS and lncRNA MIR31HG (). At the same time, no obvious heterogeneity was detected (I2 = 6.9%, PQ = 0.359), so the fixed effects model was employed. The pooled findings indicated that a shorter OS was significantly correlated with a lower lncRNA MIR31HG expression level in patients with digestive system cancers (pooled HR: 0.51; 95% CI: 0.38–0.68; A).
Table 2. Survival data of studies included in the meta-analysis.
(A) Total cancers; (B) sensitivity analysis in non-digestive system cancers.
HR: Hazard ratio.
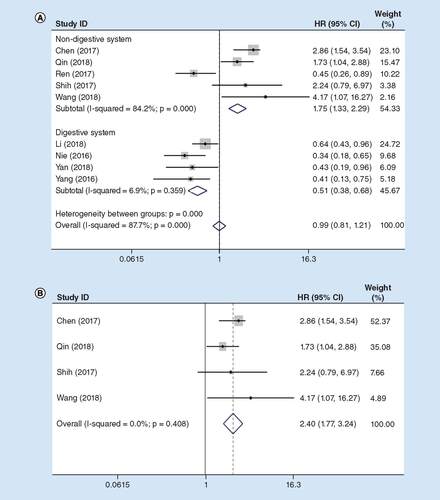
Nonetheless, the HR between high and low lncRNA MIR31HG expression groups would be 1.75 (95% CI: 1.33–2.29) in non-digestive system cancers (Figure 2B). In addition, sensitivity analysis was also carried out, and after exclusion of the study Ren et al. [Citation24], there was no heterogeneity (I2 = 0.0%, PQ = 0.408) in the non-digestive system group. Contrary to the digestive system cancer, the OS was longer in low lncRNA MIR31HG expression group of patients with non-digestive system cancer (pooled HR: 2.40; 95% CI: 1.77–3.24; B).
Therefore, lncRNA MIR31HG might serve as an indicator to independently predict the survival for cancer patients, and lncRNA MIR31HG had opposite effect on the digestive system and non-digestive system cancers.
Relationships of the lncRNA MIR31HG expression with DFS
Cumulative meta-analysis was also carried out to examine the function of lncRNA MIR31HG in DFS for 335 cancer cases enrolled from two qualified articles (Supplementary Figure 1). There was no obvious heterogeneity (I2 = 36.7%, PQ = 0.209), so the fixed effects model was used. The pooled findings suggested that low expression of lncRNA MIR31HG was related to the poor DFS (pooled HR: 0.58; 95% CI: 0.41–0.82) in digestive system cancer patients upon statistical analyses.
Relationship of the expression of lncRNA MIR31HG with LTS
illustrates the relationship of LTS with the expression of lncRNA MIR31HG based on nine articles with 796 patients. There was an obvious heterogeneity among these studies (I2 = 73.1%, PQ = 0.000), so the random-effects model was employed. According to our findings, the pooled ORs were 1.36 (95% CI: 0.59–3.14; high vs low lncRNA MIR31HG expression) in non-digestive system cancers, and 0.51 (95% CI: 0.23–1.12) in digestive system cancers (A).
(A) Total cancers; (B) sensitivity analysis in non-digestive system cancers; (C) sensitivity analysis in digestive system cancers. Weights in (A) are from random-effects analysis.
HR: Hazard ratio.
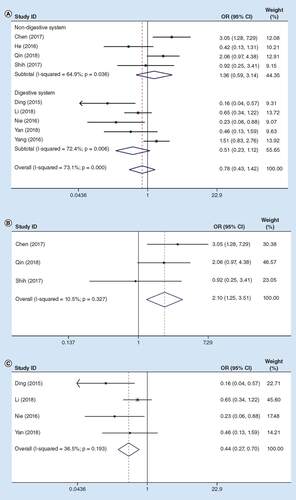
Therefore, sensitivity analysis was carried out on all the enrolled articles. According to our results, the OR between high and low expression groups was 2.10 (95% CI: 1.25–3.51) when the study by He et al. [Citation20] was eliminated (I2 = 10.5%, PQ = 0. 327) in non-digestive system cancers (B), while that was 0.44 (95% CI: 0.27–0.70) after the study by Yang et al. [Citation28] was excluded (I2 = 36.5%, PQ = 0. 193) in digestive system cancers (C).
Based on the above-mentioned results, lncRNA MIR31HG might play a distinctly different role in LTS between digestive and non-digestive systems cancers. However, difference in the LTS incidence was not statistically significant between the two groups, so more studies are warranted to verify the relationship of lncRNA MIR31HG with LTS among patients with cancer.
Relationship of the expression of lncRNA MIR31HG with PHG
Information concerning the relationship of lncRNA MIR31HG expression with PHG was extracted from all the qualified articles involving 1041 cancer patients. A significant heterogeneity was detected (I2 = 83.1%, PQ = 0.000), so the random-effects model was utilized. In addition, the OR between high and low lncRNA MIR31HG expression groups was 1.55 (95% CI: 0.56–4.30) in non-digestive system cancers, and 0.70 (95% CI: 0.27–1.79) in digestive system cancers (A).
(A) Total cancers; (B) sensitivity analysis in non-digestive system cancers; (C) sensitivity analysis in digestive system cancers. Weights in (A) and (B) are from random effects analysis.
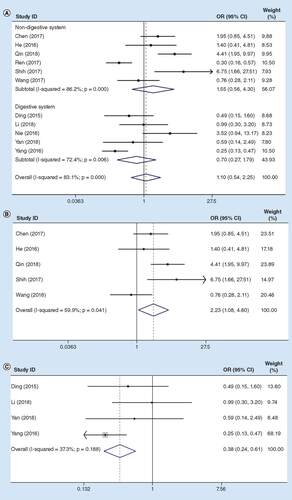
Sensitivity analysis was carried out for all the recruited articles, and after exclusion of the study Ren et al. [Citation24], the OR between high and low expression groups was 2.23 (95% CI: 1.08–4.60; B) in non-digestive system cancers, and in digestive system cancers that was 0.38 (95% CI: 0.24–0.61) after exclusion of the study Nie et al. [Citation22] (I2 = 37.3%, PQ = 0. 188; C).
Difference in the PHG incidence was statistically significant between these two groups; therefore, more studies are required to assess the association of lncRNA MIR31HG with PHG among patients with cancer.
Relationship of the expression of lncRNA MIR31HG with HTS
The correlation of HTS with the expression of lncRNA MIR31HG in this study was detected in nine eligible studies recruiting 824 patients. Our findings suggested that HTS in patients with digestive system cancers showed notable association with low expression of lncRNA MIR31HG (pooled OR: 0.29; 95% CI: 0.18–0.47; ). However, difference was not statistically significant between the high and low MIR31HG expression groups in non-digestive system cancers.
OR: Odds ratio.
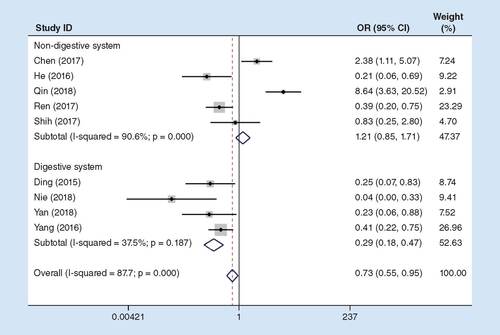
Therefore, the tumor stage in low lncRNA MIR31HG expression group was dramatically higher than that in high expression group, which demonstrated that HTS incidence showed evident correlation with the low expression of lncRNA MIR31HG in digestive system cancers.
Relationship of the expression of lncRNA MIR31HG with LNM
Information extracted based on ten qualified articles with 884 cancer patients was also examined. There was an obvious heterogeneity (I2 = 46.4%, PQ = 0.133) in digestive system cancers, so the fixed-effects model was utilized. The OR between high and low lncRNA MIR31HG expression groups was 0.58 (95% CI: 0.36–0.94; A). Consistent with the previous sensitivity analysis results, after exclusion of the study Ren et al. [Citation24], the OR between high and low expression groups was 3.25 (95% CI: 2.09–5.06) in non-digestive system cancers (I2 = 4.2%, PQ = 0.383; B).
(A) Total cancers; (B) sensitivity analysis in non-digestive system cancers.
OR: Odds ratio.
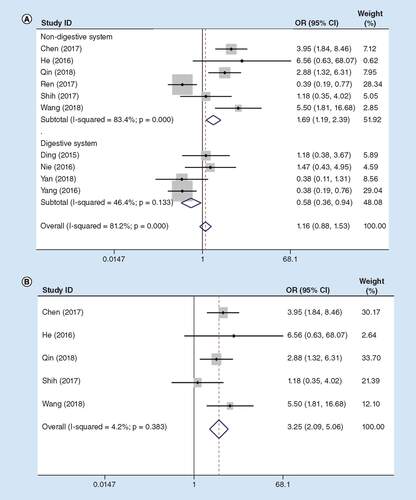
Consistent with the above findings, difference in the incidence of LNM was statistically significant between the two groups upon sensitivity analysis, so lncRNA MIR31HG might play a distinctly different role in LNM between the digestive and non-digestive systems cancers.
Relationship of the expression of lncRNA MIR31HG with DM
The relationship between DM and the expression of lncRNA MIR31HG was detected in three qualified articles with 339 patients. An obvious heterogeneity (I2 = 79.2%, PQ = 0.008), so the random effects model was utilized. The OR between high and low lncRNA MIR31HG expression groups was 1.11 (95% CI: 0.07–17.90; Supplementary Figure 2). Therefore, difference in the incidence of DM was not statistically significant between these two groups, and more studies are required to verify the relationship of lncRNA MIR31HG with DM among cancer patients.
Publication bias
Begg’s funnel plot was performed in the current meta-analysis for evaluating the potential publication bias. Supplementary Figure 3 reveals no significant asymmetry for OS (Pr>|z| = 0.754; Supplementary Figure 3A), DFS (Pr>|z| = 0.100; Supplementary Figure 3B), LTS (Pr>|z| = 0.602; Supplementary Figure 3C), PHG (Pr>|z| = 0.161; Supplementary Figure 3D), HTS (Pr>|z| = 0.251; Supplementary Figure 3E), LNM (Pr>|z| = 0.592; Supplementary Figure 3F) as well as DM (Pr>|z| = 1.000; Supplementary Figure 3G).
Discussion
Cancer remains a severe threat to human health and cancer incidence shows an increasing trend recently [Citation1]. Nonetheless, the precise metastasis mechanism in cancer patients is still unclear, even though metastasis will predict dismal prognosis [Citation29,Citation30]. Currently, identifying novel molecular markers is necessary, so as to estimate tumor metastasis, because these markers have crucial parts in cancer treatment and prediction [Citation31]. LncRNAs are one of these molecular markers, which are able to impact the development as well as occurrence of tumor; in addition, they have also been shown to display the potential of easy collection of useful biomarkers to monitor and diagnose tumors [Citation32].
LncRNA MIR31HG has been confirmed previously as a vital oncogene in a variety of human cancers, such as CVR, CRC, BC, GC, LADC, ESCC, ORC, LSCC and HCC [Citation18–Citation28]. In addition, current studies discover that lncRNA MIR31HG plays the opposite roles as a tumor-suppressor gene or an oncogene in different cancers. On the one hand, according to Nie et al., the decreased lncRNA MIR31HG expression participated in the development of GC, which might serve as a biomarker to predict the poor prognosis for GC patients [Citation22]. Yan et al. also reported that lncRNA MIR31HG positively modulated the expression of ST7L by sponging miR-575, and it played a role as a tumor suppressor in HCC [Citation27]. In CRC, MIR31HG was significantly downregulated in cancer tissues, and overexpression of MIR31HG reduced cell proliferation and induced apoptosis in vitro, and prevented tumorigenesis in vivo, while the high expression levels of LOC554202 were associated with longer DFS and OS [Citation19,Citation21,Citation28]. On the other hand, recent study confirms that high lncRNA MIR31HG expression level is an independent factor to predict the poor prognosis for LADC, and it serves an oncogene to modulate LADC cell proliferation and the cell cycle [Citation23]. Sun and his colleagues found that MIR31HG played a role as an oncogene in osteosarcoma for tumor progression and metastasis by regulating tumor-suppressor gene miR-361 and its target genes [Citation33]. MIR31HG is significantly elevated in non-small-cell lung cancer (NSCLC) tissues, and patients with higher MIR31HG expression level have a longer OS [Citation34]. Besides, Dandan et al. found that MIR31HG played a role as an oncogene role in NSCLC, and knockdown of MIR31HG expression level suppressed NSCLC cell migration, invasion and metastasis in vitro by regulating miRNA214 [Citation35]. Similar to the previous results, Wang et al. discovered that MIR31HG promoted head and neck squamous cell carcinoma (HNSCC) cell proliferation, cell cycle progression and inhibited cell apoptosis via HIF1A and p21, while MIR31HG overexpression served as a poor prognostic factor for LSCC patients [Citation26].
Moreover, most of the available literature on the relationship between lncRNA MIR31HG and cancer comes from China, but few are derived from other countries. For non-Chinese studies, Eide et al. found that aberrant expression of mir-31-5p/MIR31HG conferred intrinsic invasive and/or immunosuppressive capacity on cancer cells, and resulted in the poor prognosis for colon cancer patients [Citation36]. These results reveal that lncRNA MIR31HG serves as a vital factor to predict the prognosis for cancer patients. Nevertheless, the role of lncRNA MIR31HG in cancer remains contradictory. As a result, the current meta-analysis was carried out to explore the prognostic and clinicopathological value of lncRNA MIR31HG among cancer patients.
Information extracted from 11 qualified studies with 1041 cancer patients was analyzed in this study. Heterogeneity analysis was performed to determine whether a fixed effects model or a random-effects model should be used. In addition, differences in OS and DFS between these two groups were significant in digestive system cancer patients, when HRs based on the Cox multivariate analysis were used in combination. According to our results, the poor OS as well as DFS in digestive system cancer types was related to the low expression of lncRNA MIR31HG. In addition, the low expression of lncRNA MIR31HG in patients with digestive system cancers also displayed marked correlation with the clinicopathological variables, like HTS as well as LNM. Moreover, more studies will be required to verify the relationship of lncRNA MIR31HG with clinicopathological parameters in patients with non-digestive system cancers. Taken together, results from the current meta-analysis suggest that lncRNA MIR31HG can be used as a meaningful prognostic biomarker to predict the prognosis for numerous cancers, and lncRNA MIR31HG has opposite effect on patients with digestive system cancers compared with that on patients with non-digestive system cancers.
Limitations
Some limitations should be noted in the current meta-analysis. First, data presented in the current meta-analysis were not applicable to all countries worldwide, because all of them were derived from China. Second, only 11 studies were finally collected into the current study regardless of the tremendous efforts in literature retrieval, and our conclusions might become invalid due to the small sample size. Third, there was no identical standard of high expression among the enrolled studies, and the same value could hardly be obtained. Finally, some confounding factors might impact the prognosis for cancer patients, like co-morbidity as well as treatment; however, there were no available data from the recruited studies. Thus, it was the intrinsic drawback of the current systematic review as well as meta-analysis. Therefore, further well-designed high-quality studies are warranted, to examine the effect of lncRNA MIR31HG on cancer.
Conclusion
Taken together, this study indicates that the low expression of lncRNA MIR31HG in digestive system cancer is significantly related to the dismal OS, DFS, HTS as well as LNM. Therefore, lncRNA MIR31HG is promising to be used as a predicting factor for the metastasis as well as prognosis in patients with digestive system cancer.
Future perspective
LncRNA MIR31HG is a potentially suitable clinicopathological and prognostic biomarker in digestive system cancers. Nevertheless, more well-designed studies with high quality are required to examine the effect of lncRNA MIR31HG on cancer, especially for non-digestive cancers. Additionally, relevant research from more countries is needed to make the results more reliable.
Background
Abnormal expression of long noncoding RNA MIR31 host gene (lncRNA MIR31HG) can be frequently detected in cancer.
LncRNA MIR31HG may affect the metastasis and prognosis for cancer, however, most current research is restricted due to the small sample size and the discrete outcomes.
We performed a meta-analysis to examine the significance of lncRNA MIR31HG in predicting the metastasis and prognosis for patients with cancer.
Materials & methods
Web of Science database, PubMed and OVID were used to search available studies.
Statistical analysis was performed using the Stata12.0 software.
Results
Low expression of lncRNA MIR31HG in digestive system cancer was significantly related to the dismal overall survival, disease-free survival, high tumor stage and lymph node metastasis.
Discussion
LncRNA MIR31HG is promising to be used as a predicting factor for the metastasis and prognosis for patients with digestive system cancer.
Supplementary file
Download Zip (2.9 MB)Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.tandfonline.com/doi/suppl/10.2217/bmm-2019-0145
Financial & competing interests disclosure
This manuscript was funded by the Science and Technology Development Program of Shandong Province (no. 2015GSF118110), and the Graduate Innovation Fund of Peking Union Medical College (2018-1002-01-10). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- Siegel RL , MillerKD, JemalA. Cancer statistics, 2019. CA Cancer J. Clin.69(1), 7–34 (2019).
- Fan YH , FangH, JiCX, XieH, XiaoB, ZhuXG. Long noncoding RNA CCAT2 can predict metastasis and poor prognosis: a meta-analysis. Clin. Chim. Acta466, 120–126 (2017).
- Jiang MC , NiJJ, CuiWY, WangBY, ZhuoW. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am. J. Cancer Res.9(7), 1354–1366 (2019).
- Zheng Y , GaoY, LiXet al. Long non-coding RNA NAP1L6 promotes tumor progression and predicts poor prognosis in prostate cancer by targeting Inhibin-beta A. Onco. Targets Ther.11, 4965–4977 (2018).
- Hua JT , AhmedM, GuoHet al. Risk SNP-mediated promoter-enhancer switching drives prostate cancer through lncRNA PCAT19. Cell174(3), 564e518–575e518 (2018).
- Sun L , WangL, ChenTet al. microRNA-1914, which is regulated by lncRNA DUXAP10, inhibits cell proliferation by targeting the GPR39-mediated PI3K/AKT/mTOR pathway in HCC. J. Cell. Mol. Med. doi:10.1111/jcmm.14705 (2019) ( Epub ahead of print).
- Xu Y , LiY, JinJet al. LncRNA PVT1 up-regulation is a poor prognosticator and serves as a therapeutic target in esophageal adenocarcinoma. Mol. Cancer18(1), 141 (2019).
- Fan Y , YanT, ChaiY, JiangY, ZhuX. Long noncoding RNA HOTTIP as an independent prognostic marker in cancer. Clin. Chim. Acta482, 224–230 (2018).
- Yousefi H , MaheronnaghshM, MolaeiFet al. Long noncoding RNAs and exosomal lncRNAs: classification, and mechanisms in breast cancer metastasis and drug resistance. Oncogene doi:10.1038/s41388-019-1040-y (2019) ( Epub ahead of print).
- Fan Y , HeY, ZhouX, LiuY, WangF. Meta-analysis of the prognostic value of lncRNA DANCR for cancer patients in China. Cancer Manag. Res.11, 2027–2037 (2019).
- Cai P , LiH, HuoWet al. Aberrant expression of LncRNA-MIR31HG regulates cell migration and proliferation by affecting miR-31 and miR-31* in Hirschsprung's disease. J. Cell. Biochem.119(10), 8195–8203 (2018).
- Gao J , ChenF, HuaMet al. Knockdown of lncRNA MIR31HG inhibits cell proliferation in human HaCaT keratinocytes. Biol. Res.51(1), 30 (2018).
- Montes M , NielsenMM, MaglieriGet al. The lncRNA MIR31HG regulates p16(INK4A) expression to modulate senescence. Nat. Commun.6, 6967 (2015).
- Altman DG , McshaneLM, SauerbreiW, TaubeSE. REporting recommendations for tumor MARKer prognostic studies (REMARK): explanation and elaboration. PLoS Med.9(5), e1001216 (2012).
- Harris AL . REporting recommendations for tumour MARKer prognostic studies (REMARK). Br. J. Cancer93(4), 385–386 (2005).
- Fan YH , JiCX, XuB, FanHY, ChengZJ, ZhuXG. Long noncoding RNA activated by TGF-beta in human cancers: a meta-analysis. Clin. Chim. Acta468, 10–16 (2017).
- Mei H , LiuY, ZhouQ, HuK, LiuY. Long noncoding RNA MALAT1 acts as a potential biomarker in cancer diagnosis and detection: a meta-analysis. Biomark. Med.13(1), 45–54 (2019).
- Chen J , ZhuJ. Elevated expression levels of long non-coding RNA, Loc554202, are predictive of poor prognosis in cervical cancer. Tohoku J. Exp. Med.243(3), 165–172 (2017).
- Ding J , LuB, WangJet al. Long non-coding RNA Loc554202 induces apoptosis in colorectal cancer cells via the caspase cleavage cascades. J. Exp. Clin. Cancer Res.34, 100 (2015).
- He A , ChenZ, MeiH, LiuY. Decreased expression of LncRNA MIR31HG in human bladder cancer. Cancer Biomark.17(2), 231–236 (2016).
- Li Y , XinS, WuHet al. High expression of microRNA31 and its host gene LOC554202 predict favorable outcomes in patients with colorectal cancer treated with oxaliplatin. Oncol. Rep.40(3), 1706–1724 (2018).
- Nie FQ , MaS, XieM, LiuYW, DeW, LiuXH. Decreased long noncoding RNA MIR31HG is correlated with poor prognosis and contributes to cell proliferation in gastric cancer. Tumour Biol.37(6), 7693–7701 (2016).
- Qin J , NingH, ZhouY, HuY, YangL, HuangR. LncRNA MIR31HG overexpression serves as poor prognostic biomarker and promotes cells proliferation in lung adenocarcinoma. Biomed. Pharmacother.99, 363–368 (2018).
- Ren ZP , ChuXY, XueZQet al. Down-regulation of lncRNA MIR31HG correlated with aggressive clinicopathological features and unfavorable prognosis in esophageal squamous cell carcinoma. Eur. Rev. Med. Pharmacol. Sci.21(17), 3866–3870 (2017).
- Shih JW , ChiangWF, WuATHet al. Long noncoding RNA LncHIFCAR/MIR31HG is a HIF-1alpha co-activator driving oral cancer progression. Nat. Commun.8, 15874 (2017).
- Wang R , MaZ, FengLet al. LncRNA MIR31HG targets HIF1A and P21 to facilitate head and neck cancer cell proliferation and tumorigenesis by promoting cell-cycle progression. Mol. Cancer17(1), 162 (2018).
- Yan S , TangZ, ChenKet al. Long noncoding RNA MIR31HG inhibits hepatocellular carcinoma proliferation and metastasis by sponging microRNA-575 to modulate ST7L expression. J. Exp. Clin. Cancer Res.37(1), 214 (2018).
- Yang L , WeiH, XiaoHJ. Long non-coding RNA Loc554202 expression as a prognostic factor in patients with colorectal cancer. Eur. Rev. Med. Pharmacol. Sci.20(20), 4243–4247 (2016).
- Suhail Y , CainMP, VanajaKet al. Systems biology of cancer metastasis. Cell Syst.9(2), 109–127 (2019).
- Chang LC , FanCW, TsengWKet al. The ratio of thioredoxin/Keap1 protein level is a predictor of distant metastasis in colorectal cancer. Biomark. Med.11(12), 1103–1111 (2017).
- Zhou X , FanYH, WangY, WangF, LiuY. Prognostic value of long non-coding RNA ZEB1-AS1 in Chinese cancer patients: A Meta-analysis. Medicine (Baltimore)98(17), e15251 (2019).
- Qi P , DuX. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod. Pathol.26(2), 155–165 (2013).
- Sun Y , JiaX, WangM, DengY. Long noncoding RNA MIR31HG abrogates the availability of tumor suppressor microRNA-361 for the growth of osteosarcoma. Cancer Manag. Res.11, 8055–8064 (2019).
- Zheng S , ZhangX, WangX, LiJ. MIR31HG promotes cell proliferation and invasion by activating the Wnt/beta-catenin signaling pathway in non-small cell lung cancer. Oncol. Lett.17(1), 221–229 (2019).
- Dandan W , JianliangC, HaiyanH, HangM, XuedongL. Long noncoding RNA MIR31HG is activated by SP1 and promotes cell migration and invasion by sponging miR-214 in NSCLC. Gene692, 223–230 (2019).
- Eide PW , EilertsenIA, SveenA, LotheRA. Long noncoding RNA MIR31HG is a bona fide prognostic marker with colorectal cancer cell-intrinsic properties. Int. J. Cancer144(11), 2843–2853 (2019).