Epigenetics and epigenomics have taken center stage for many questions in biomedical research in the last decade. They have moved from a scientific discipline restricted to a limited number of expert laboratories to a commonly analyzed molecular level accessible to any laboratory performing molecular biology. Epigenetics has been omnipresent over the last decade and epigenetics and epigenomics have experienced a lot of hype. Epigenetics was regarded as a solution for resolving the missing heritability, coming with the disappointing realization that large-scale genetic studies, particularly genome-wide association studies (GWAS), did not fulfill the initial promise of explaining complex diseases. In fact, with the disappointment in genetics, epigenetics was seen as the solution to everything; with its aura of mystery, it was the one-word solution to every question, leading to the attitude of “if you can not explain it with your genomic data, then it is probably epigenetics”.
Epigenomics has been with the community for the last 10 years and has been providing a publication platform with a well-balanced mixture of novel research results mainly related to human diseases, thought provoking comments and comprehensive reviews, which allowed readers to keep up-to-date in this rapidly moving field.
Epigenetic research has been largely driven by technological progress, notably high-throughput sequencing technologies, which allow us to analyze multiple epigenetic levels at single-nucleotide resolution on the same read-out platform, facilitating data integration. While the epigenetic components, associated DNA methylation changes and chromatin reorganization in cancer have been known for quite some time, the last decade has demonstrated (in a multitude of studies) that changes are also present in a large variety of complex diseases, including autoimmune, inflammatory, neurodegenerative and metabolic disorders. In analogy to the analysis of genetic variation in GWAS, the last decade has seen the emergence of epigenome-wide association studies (EWAS), which commonly use BeadArray-based technologies to interrogate associations of between 2 and 3% of the CpGs in the human genome with a phenotype of interest. Some of the first EWAS published were statistically underpowered and confounded by nonphenotype-related differences such as differences in age or sample composition. Study design and statistical methods have since greatly improved. For the analysis and detection of potential confounding factors, current EWAS do yield changes that can be reproduced in different cohorts, albeit with some room for improvement.
Methods analyzing locus-specific DNA methylation patterns have been shown to be mature and reproducible, allowing the translation from a research laboratory-based test to the analysis in clinical routine. The first commercial DNA methylation-based biomarkers have now been approved by American, Chinese and European regulatory agencies and have been rolled out, especially in the field of screening at-risk populations for the development of colorectal cancer.
Circulating cell-free DNA and nucleosomes released from dying cells have emerged as a new class of biomarkers due to the cell-type specific patterns of epigenetic modification which allow us to retrace the origin of the dying cells, thus determining the origin of cancers, while also assisting in monitoring patients’ responses to therapy. While pioneered in the field of cancer, applications have diversified to include rejection of transplants, traumatic brain injury and detection of diabetes.
The articles published in Epigenomics reflect these evolutions and the maturation of the field. shows the ten most cited articles published so far in Epigenomics, while provides an overview of the 20 most trending and influential articles published in the last 10 years in Epigenomics. These include widely read review articles, the evaluation of novel technologies, creation of computational tools rapidly adopted by the community and primary research articles shedding new light on specific research topics.
Table 1. The ten most cited articles published in Epigenomics.
Table 2. The 20 most trending articles published in Epigenomics.
In line with the importance of technological evolution, four out of the ten most cited papers review, evaluate or provide novel analysis tools for the widely used epigenotyping BeadChips, including the first report on the performance of the current generation, the EPIC BeadChips. The top ten articles also include descriptions of other useful data resources, as well as reviews on how diet and other exposures shape the epigenome, with two review articles describing aspects of epigenetic changes in cancer. Our top cited article has received more than 400 citations reviewing the role of hypomethylation in cancer.
The most influential articles as measured by the Altmetric attention score () comprise different levels of gene regulation, including DNA methylation, histone modification and small RNAs such as microRNAs. Of note, most of the attention-receiving articles were on subjects outside the cancer field. While some technology reports and computational tools are found in both categories, many of these influential articles address less well-known (and perhaps more controversial) aspects of epigenetics, including the impact of exercise, socioeconomic status or social stress on epigenetically mediated gene regulation: epigenetic changes drawing a connection between our daily life and epigenetics. Their high attention score demonstrates that these topics are of interest to the much broader public.
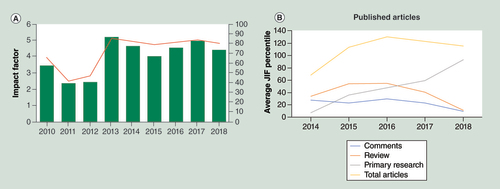
Epigenomics has rapidly been adopted as a publication platform by the research community and has established itself among the leading journals in epigenetic research (A) and among the top 20% journals in genomics and genetics. Over the years, the proportion of primary research has greatly increased and represents now 79.6% of the accepted manuscripts (B). High quality of the published articles is ensured through thorough but constructive peer review with an acceptance rate for submitted articles of 36.8% in 2018. The success of Epigenomics is also reflected in the increase in the volumes per year. While until 2014 Epigenomics published six issues per year, the number was gradually increased and 115 articles were published in 16 issues in 2018. This trend is set to continue with the issues increasing to 24 in 2020.
(A) Performance of the impact factor (left y axis) as well as the journal impact factor percentiles (right y axis) allowing cross journal comparison for Epigenomics. Data from InCites Journal Citation Reports dataset updated 20 September 2019. (B) Evolution of accepted submissions in Epigenomics showing the strong increase in primary research articles over the last 4 years. The commentary category also includes editorials, conference reports and interviews with leading experts in the field.
JIF: Journal impact factor.
The last decade has been marked by the increased possibilities of genetic editing using mainly the CRISPR/Cas9 system. Epigenetic editing follows a similar approach, but instead of cutting the genome, an epigenetic modifier is targeted to a specific locus, where it changes the epigenetic landscape to open up or close the locus for transcription. Clinical applications, especially in diseases without strong genetic drivers (e.g. mutations), might be possible in the future; this strategy provides, for the first time, the tools to investigate relatively easily the functional relevance of epigenetic changes.
However, much remains to be done. Many of the large cohorts which have so far been analyzed have been collected for genetic analyses. Although we have learned to infer proportions and correct for biases in the composition of major blood cell populations, for many phenotypes blood might not be the most informative tissue to analyze and establishment of cell-type specific reference maps for other tissues is associated with significant efforts. In the same line of thought, sampling from clinical trials rarely includes samples dedicated to the analysis of epigenetic changes from the design of the study. The investigation of epigenetic changes to a priori identify responders before the initiation of treatment and understand epigenetic contributions to the mechanisms of drug resistance bear great potential to improve patient management in the future.
Although technologies have been greatly improved and have been largely responsible for the advances of the field, several challenges remain. Alternatives to bisulfite treatment (which substantially degrades the DNA) using alternative chemistries or direct read-out of epigenetic modifications have been devised, but are only just starting to be adopted by a wider community. While the feasibility of single cell epigenomics has been shown, improvements on the amplification steps will be required to allow a more robust, comprehensive and potentially multi-level characterization of the epigenome of single cells. Application of the approach of spatial transcriptomics, which allows the measurement of gene expression patterns of cells in their microenvironment, would also be of great interest for the analysis of epigenetic modifications.
There are exciting times before us and Epigenomics will continue to provide the epigenetic community with a high-quality forum to widely disseminate the results, which will help to improve our understanding of the complex gene-regulatory networks set up by epigenetic modifications and their alterations in disease. This will lead to improved and more tailored patient management in the future.
I am indebted to the hard-working staff at Future Medicine handling the manuscripts on a day-by-day basis and to our expert Editorial Board, who among their multiple engagements find the time to review manuscripts and provide valuable input on the overall strategy of the journal. But most all, my thanks go to you, our readers and past or future contributors.
We are looking forward to your submissions to continue this journey.
Jorg Tost.
Financial & competing interests disclosure
The author has no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
- Ehrlich M . DNA hypomethylation in cancer cells. Epigenomics1(2), 239–259 (2009).
- Bibikova M , LeJ , BarnesBet al. Genome-wide DNA methylation profiling using Infinium® assay. Epigenomics1(1), 177–200 (2009).
- Dedeurwaerder S , DefranceM , CalonneE , DenisH , SotiriouC , FuksF. Evaluation of the Infinium Methylation 450K technology. Epigenomics3(6), 771–784 (2011).
- Touleimat N , TostJ. Complete pipeline for Infinium® Human Methylation 450K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics4(3), 325–341 (2012).
- Reichard JF , PugaA. Effects of arsenic exposure on DNA methylation and epigenetic gene regulation. Epigenomics2(1), 87–104 (2010).
- Alegría-Torres JA , BaccarelliA , BollatiV. Epigenetics and lifestyle. Epigenomics3(3), 267–277 (2011).
- Hardy TM , TollefsbolTO. Epigenetic diet: impact on the epigenome and cancer. Epigenomics3(4), 503–518 (2011).
- Moran S , ArribasC , EstellerM. Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics8(3), 389–399 (2016).
- Hashimoto H , VertinoPM , ChengX. Molecular coupling of DNA methylation and histone methylation. Epigenomics2(5), 657–669 (2010).
- Chadwick LH . The NIH Roadmap Epigenomics Program data resource. Epigenomics4(3), 317–324 (2012).
- Denham J , O’BrienBJ , HarveyJT , CharcharFJ. Genome-wide sperm DNA methylation changes after 3 months of exercise training in humans. Epigenomics7(5), 717–731 (2015).
- McCarthy D , PulvererW , WeinhaeuselAet al. MethylMeter®: bisulfite-free quantitative and sensitive DNA methylation profiling and mutation detection in FFPE samples. Epigenomics8(6), 747–765 (2016).
- Liang L , LutzBM , BekkerA , TaoY-X. Epigenetic regulation of chronic pain. Epigenomics7(2), 235–245 (2015).
- Moody L , ChenH , PanYX. Postnatal diet remodels hepatic DNA methylation in metabolic pathways established by a maternal high-fat diet. Epigenomics9(11), 1387–1402 (2017).
- Roy R , ChatterjeeA , DasDet al. Genome-wide miRNA methylome analysis in oral cancer: possible biomarkers associated with patient survival. Epigenomics11(5), 473–487 (2019).
- Calcagno DQ , WisnieskiF , DaSilva Mota ERet al. Role of histone acetylation in gastric cancer: implications of dietetic compounds and clinical perspectives. Epigenomics11(3), 349–362 (2019).
- Armstrong NJ , MatherKA , ThalamuthuAet al. Aging, exceptional longevity and comparisons of the Hannum and Horvath epigenetic clocks. Epigenomics9(5), 689–700 (2017).
- Moran S , ArribasC , EstellerM. Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics8(3), 389–399 (2016).
- De Vega WC , ErdmanL , VernonSD , GoldenbergA , McGowanPO. Integration of DNA methylation & health scores identifies subtypes in myalgic encephalomyelitis/chronic fatigue syndrome. Epigenomics10(5), 539–557 (2018).
- Bush NR , EdgarRD , ParkMet al. The biological embedding of early-life socioeconomic status and family adversity in children’s genome-wide DNA methylation. Epigenomics10(11), 1445–1461 (2018).
- LaSalle JM , YasuiDH. Evolving role of MeCP2 in Rett syndrome and autism. Epigenomics1(1), 119–130 (2009).
- Cunliffe VT . The epigenetic impacts of social stress: how does social adversity become biologically embedded?Epigenomics8(12), 1653–1669 (2016).
- Sarkar S , GoldgarS , BylerS , RosenthalS , HeerbothS. Demethylation and re-expression of epigenetically silenced tumor suppressor genes: sensitization of cancer cells by combination therapy. Epigenomics5(1), 87–94 (2013).
- Rosato AJ , ChenX , TanakaYet al. Salivary microRNAs identified by small RNA sequencing and machine learning as potential biomarkers of alcohol dependence. Epigenomics11(7), 739–749 (2019).
- Chatterjee A , StockwellPA , RodgerEJ , ParryMF , EcclesMR. Scan-tcga tools for integrated epigenomic and transcriptomic analysis of tumor subgroups. Epigenomics8(10), 1315–1330 (2016).
- Sánchez-Conde M , Rodriguez-CentenoJ , DrondaFet al. Frailty phenotype: a clinical marker of age acceleration in the older HIV-infected population. Epigenomics11(5), 501–509 (2019).
- Chu SH , LoucksEB , KelseyKTet al. Sex-specific epigenetic mediators between early life social disadvantage and adulthood BMI. Epigenomics10(6), 707–722 (2018).
