Welcome to the 13th volume of Epigenomics. I would like to take this opportunity to wish our readers a happy new year. In this short Foreword, I will be looking back at some of the journal highlights of 2020. 2020 has presented unprecedented challenges for all, our authors and readership included. Despite laboratory closures, adjustments to work and varying personal circumstances, authors for Epigenomics have continued to advance the development of epigenetics for medical applications with research and reviews of the highest quality, which it has been our privilege to publish.
Article highlights
Epigenomics has published several exciting articles this year, some of which I would like to draw your attention to here ().
Most read articles
At the time of writing (November 2020) the most read research article published in 2020 was ‘Multi-omics analysis based on integrated genomics, epigenomics and transcriptomics in pancreatic cancer’ by Kong and co-authors from Shengjing Hospital of China Medical University (Shengyang, China) and Ludwig-Maximilians-University Munich (Munich, Germany) [Citation1]. By analysis of data from The Cancer Genome Atlas, the authors identified four molecular subgroups of pancreatic cancer that may help in the discovery of new prognostic biomarkers for one of the leading causes of cancer-related death.
Another of our top-accessed articles this year highlighted the importance of DNA methylation markers in epigenetics for cancer diagnostics. In ‘Genome-wide analysis of DNA methylation identifies two CpG sites for the early screening of colorectal cancer’ by Wang and co-authors [Citation2], the authors used data from 15 cancer types to construct a CpG-methylation-based model that could be a powerful tool for colorectal cancer screening. The next most read article, ‘A risk prediction model of DNA methylation improves prognosis evaluation and indicates gene targets in prostate cancer’ by Zhang and co-authors used DNA methylation data to develop a risk assessment system, which in functional validation revealed an important gene target for prostate cancer [Citation3].
In addition to research, other top-accessed content included our 10-year anniversary Foreword from Senior Editor, Jörg Tost, CEA-Institut de Biologie François Jacob (France) [Citation4], a review on the role of circular RNA in mediating resistance to anticarcinogens from Geng and co-authors [Citation5] and an exciting Editorial about the newly launched Physiome Project from Noble and Hunter [Citation6].
Table 1. Top-accessed articles in 2020.
Articles about SARS-CoV-2 during the COVID-19 pandemic
Not only did contributions to the journal continue during the COVID-19 pandemic, but Epigenomics was able to publish a number of articles to support efforts to tackle the virus. In ‘Targeting SARS-CoV-2 using polycomb inhibitors as antiviral agents’, authors Crea and Ayaz discussed the potential to harness the epigenetic target EZH2 to treat patients with COVID-19 [Citation7]. ‘Potential use of noncoding RNAs and innovative therapeutic strategies to target the 5′UTR of SARS-CoV-2’ by Baldassarre and co-authors offered a forward-looking take on the strategies to target specific regions of the virus’ genome [Citation8]. Li and co-authors contributed their research entitled ‘An integrative analysis identifying transcriptional features and key genes involved in COVID-19’, which identified key players in infection by the novel coronavirus, to support therapeutic development [Citation9].
Impact&reach of Epigenomics
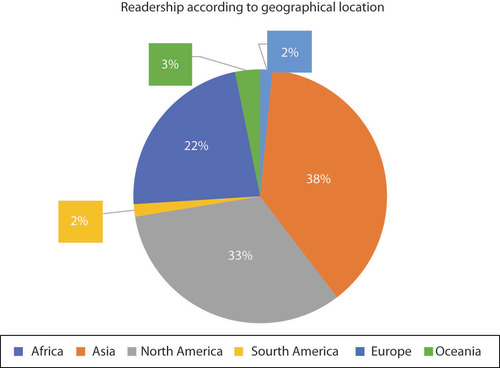
Audience locations
Epigenomics remains a globally accessed publication. In 2020, we were pleased to see our audiences in North America, Asia and Europe remain widely engaged while promising growth in our African readership begins to more appropriately reflect the vital research community there (). This year, Future Science Group has updated its policy on helicopter research in order to ensure our publications properly reflect the contributions of researchers worldwide.
Impactful articles in the news&social media
Altmetrics provide insights into the impact of an article by calculating an ‘attention score’ based on mentions from social media and news outlets [Citation10]. The highest Altmetrics attention score in 2020 was for ‘Transcriptomic and epigenomics atlas of myotubes reveals insight into the circadian control of metabolism and development’ by Altıntaş and co-authors, which gained an attention score of 20 for its high number of mentions on Twitter [Citation11]. ‘Comparison of smoking-related DNA methylation between newborns from prenatal exposure and adults from personal smoking’, which was published in 2019, gained attention early in 2020 when it was highlighted in the NIH ‘I am Intramural Blog’ for its insights into the epigenetic effects of smoking while pregnant [Citation12,Citation13]. Epigenomics has a Twitter account, which is a great way to hear about the latest news and articles from the journal. Follow us @fsgepi or join the Epigenomics LinkedIn group for more updates from Future Science Group [Citation14,Citation15].
Conclusion
We are grateful to the authors, reviewers and readers of Epigenomics, who are vital to the continued growth and reach of the journal. Despite the challenges of 2020, our contributors have remained committed to the publication of high-quality articles that advance the field of epigenetics for medical application. With your contributions, the journal remains at the forefront of publishing in its field and we look forward to publishing further valuable research, review and opinion articles in the coming year.
Epigenomics gladly considers unsolicited articles, including original research, reviews, editorials and more. More information on article types and submitting to the journal can be found on our website.
Financial&competing interest disclosure
C Brady is an employee of Future Medicine Ltd, publisher of Epigenomics. The author has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- Kong L , LiuP , ZhengM , XueB , LiangK , TanX. Multi-omics analysis based on integrated genomics, epigenomics and transcriptomics in pancreatic cancer. Epigenomics12(6), 507–524 (2020).
- Wang X , WangD , ZhangH , FengM , WuX. Genome-wide analysis of DNA methylation identifies two CpG sites for the early screening of colorectal cancer. Epigenomics12(1), 37–52 (2020).
- Zhang E , HouX , HouB , ZhangM , SongY. A risk prediction model of DNA methylation improves prognosis evaluation and indicates gene targets in prostate cancer. Epigenomics12(4), 333–352 (2020).
- Tost J . 10 years of Epigenomics: a journey with the epigenetic community through exciting times. Epigenomics12(2), 81–85 (2020).
- Geng X , JiaY , ZhangYet al. Circular RNA: biogenesis, degradation, functions and potential roles in mediating resistance to anticarcinogens. Epigenomics12(3), 267–283 (2020).
- Noble D , HunterP. How to link genomics to physiology through epigenomics. Epigenomics12(4), 285–287 (2020).
- Ayaz S , CreaF. Targeting SARS-CoV-2 using polycomb inhibitors as antiviral agents. Epigenomics12(10), 811–812 (2020).
- Baldassarre A , PaoliniA , BrunoSP , FelliC , TozziAE , MasottiA. Potential use of noncoding RNAs and innovative therapeutic strategies to target the 5′UTR of SARS-CoV-2. Epigenomics12(15), 1349–1361 (2020).
- Li G , WangJ , HeXet al. An integrative analysis identifying transcriptional features and key genes involved in COVID-19. Epigenomics12(22), 1969–1981 (2020).
- Discover the attention surrounding your research – Altmetric. www.altmetric.com/
- Altıntaş A , LakerRC , GardeC , BarrèsR , ZierathJR. Transcriptomic and epigenomics atlas of myotubes reveals insight into the circadian control of metabolism and development. Epigenomics12(8), 701–713 (2020).
- Sikdar S , JoehanesR , JoubertBRet al. Comparison of smoking-related DNA methylation between newborns from prenatal exposure and adults from personal smoking. Epigenomics11(13), 1487–1500 (2019).
- Mothers’ smoking leaves unique marks on infants’ DNA | NIH intramural research program. www.irp.nih.gov/blog/post/2020/01/mothers-smoking-leaves-unique-marks-on-infants-dna
- Epigenomics Journal (@fsgepi) / Twitter. www.twitter.com/fsgepi
- Epigenomics | Groups | LinkedIn . www.linkedin.com/groups/8267879/