Abstract
Aim: To explore the role of RanBP9 in breast cancer. Materials & methods: Oncomine, TIMER, GEPIA, UALCAN, c-BioPortal databases and tissue microarray analysis were used in this study. Results: The expression level of RanBP9 is elevated in breast cancer tissues, which is associated with poor prognosis in breast cancer patients. RanBP9 exhibits genetic alterations and a decreased methylation level in cancer tissues. RanBP9 may also regulate cell cycle progression and is linked to tumor purity and the infiltrating levels of immune cells. Conclusions:RanBP9 may correlate with prognosis and immune infiltration in breast cancer, laying the foundation for future studies on the potential role of RanBP9 in breast cancer.
Lay abstract
RanBP9 has diverse function in various tumor types. However, the role of RanBP9 in breast cancer remains to be explored. In this study, we investigated the gene expression of RanBP9 using databases and clinical samples. We found the expression level of RanBP9 is raised in breast cancer tissues and is associated with poor prognosis for breast cancer patients. RanBP9 may be involved in the regulation of the cell cycle and related to tumor immunity. Overall, we found that RanBP9 may correlate with prognosis and immune infiltration of breast cancer, laying the foundation for future studies on the potential role of RanBP9 in breast cancer.
In 2020, the incidence rate of breast cancer in women exceeded lung cancer and became the most prevalent cancer type in the world with 2,300,000 new cases estimated. For women, breast cancer is not only the most common cancer but also the leading cause of cancer death [Citation1]. Risk factors for breast cancer can be genetic and nongenetic. Genetic factors include mutations in susceptible genes – mainly BRCA1, BRCA2 or CHEK2 – and SNPs. Nongenetic factors include reproductive and hormonal risk factors such as early menarche age, high birth age, shorter breastfeeding periods, late menopause, menopausal hormone therapy, high mammographic density, chest radiation exposure, insufficient exercise, alcohol and obesity [Citation2]. On the basis of the behaviors of ER, PR, HER-2 and Ki-67, breast cancer can be classified into four types: luminal A, luminal B, HER-2 overexpression and basal-like [Citation3,Citation4]. With the development of molecular biology, bioinformatics and various omics technologies, more accurate classification can be carried out on the basis of quadruple classification. In addition to the classic markers ER, PR and HER-2, more biomarkers need to be discovered to assist the diagnosis, molecular typing, treatment and prognosis of breast cancer. Currently, the diagnosis and treatment of breast cancer has used a molecular typing model, which greatly improves the prognosis of breast cancer patients [Citation5–7]. It is expected that the treatment of breast cancer will enter a precise and individualized treatment stage. However, the heterogeneity of cancer cells remains an obstacle to finding a cure.
RanBP9 (also called RanBPM), with a molecular weight of 90 kDa, is localized in the nucleus and cytoplasm of a cell. It is highly conserved in various organisms and widely expressed in different tissue types [Citation8]. RanBP9 is a member of a macromolecular complex termed carboxy-terminal to LisH (CTLH), which has 11 members, with the remaining ten members including ARMC8, GID4, GID8, MAEA, MKLN1, RMND5A, RMND5B, RanBP10, WDR26 and YPEL5. CTLH protein is linked to many basic biological processes, including proliferation, cell adhesion, migration, survival and programmed cell death [Citation9,Citation10]. Recent studies have shown that as a member of the CTLH, RanBP9 is involved in the regulation of various pathways and cellular processes, including cell apoptosis, transcriptional regulation, cell adhesion and migration, cellular morphology, ubiquitination and DNA damage repair [Citation11–13]. A role of RanBP9 in cancer has been documented, but the findings are conflictive. RanBP9 has a dual effect on tumor progression. On one hand, it is manifested as a tumor suppressor. For instance, RanBP9 inhibits the proliferation and invasion of lung cancer, gastric cancer and colon cancer cells [Citation14–16]. On the other hand, it was also reported that RanBP9 plays a role in promoting cancerous behaviors, such as the migration of renal cancer cells [Citation17,Citation18]. Although the results from in vitro experiments show that knockdown of RanBP9 leads to increased proliferation of tumor cells, the level of RanBP9 transcripts in tumor samples is higher than that in nontumor samples [Citation19]. For example, RanBP9 exhibits higher levels of expression in lung cancer, colorectal cancer, osteosarcoma gastric cancer and invasive breast cancer [Citation9]. In addition, studies have shown that RanBP9 deletion can increase the sensitivity of lung cancer to the treatment of genotoxicity, indicating that RanBP9 may be a potential therapeutic target [Citation20].
In this study, we used data mining to study the relationship between RanBP9 and breast cancer, including its expression and its association with prognosis, genetic changes, functional networks, immune cell infiltration and protein interactions. The findings reported here indicate that the expression of RanBP9 in breast cancer tissues is elevated as verified in tissue microarray and that the expression of RanBP9 may be linked to the prognosis and immune infiltration of breast cancer, thereby laying the foundation for future studies on the role of RanBP9 in the diagnosis, prognosis and treatment of breast cancer. Our studies also implicate RanBP9 a potential marker and therapeutic target for breast cancer.
Materials & methods
Oncomine analysis
Oncomine (www.oncomine.org) is currently the world’s largest online cancer microarray database, including more than 80,000 cancer and normal tissues [Citation21]. The transcription levels of RanBP9 in different types of cancer and corresponding normal tissues were compared using the threshold of p-value ≤0.05, fold-change ≥2.
UALCAN analysis
UALCAN (http://ualcan.path.uab.edu) is an interactive web portal for in-depth analysis of The Cancer Genome Atlas (TCGA) gene expression data which uses TCGA Level 3 RNA SEQ and clinical data from 31 cancer types [Citation22]. It can not only analyze the expression level of specific genes in different cancer and normal tissues but also define the subsets to query the relative expression of genes in tumor and normal samples based on clinicopathological characteristics such as gender, race and tumor grade. Subgroup analysis can help better understand specific diseases. In addition, this tool was also used to analyze differences in promoter methylation levels.
GEPIA dataset
GEPIA (gene expression profiling interactive analysis) provides key interactive and customizable functions based on data from TCGA and the Genotype-Tissue Expression (GTEx) projects, including differential expression analysis, spectrum drawing, correlation analysis and patient survival analysis, for example [Citation23]. We used this tool to analyze the expression of RanBP9 in breast cancer. GEPIA is available at http://gepia.cancer-pku.cn/.
The Kaplan–Meier Plotter
Kaplan–Meier Plotter (www.kmplot.com) is an online database containing gene expression data and clinical data, mainly of lung, ovarian, gastric and breast cancers. To evaluate the prognostic value of specific genes, patient samples were divided into two groups based on the gene expression level (high expression and low expression). By automatically selecting the best cutoff value to divide breast cancer patients, the relationship between RanBP9 expression and prognosis was analyzed with the hazard ratio (HR) with 95% CIs and log rank p-value. The prognostic indicators evaluated include relapse-free survival (RFS), overall survival (OS), post-progression survival (PPS) and distant-metastasis-free survival (DMFS). The relationship between RanBP9 expression and the prognosis of different subtypes of breast cancer was also analyzed. The prognostic index is for RFS, whereas other conditions were as described earler.
Tumor Immune Estimation Resource
Tumor Immune Estimation Resource (TIMER; cistrome.shinyapps.io/timer) collects 10,897 samples of 32 cancer types from TCGA, providing a tool for a comprehensive study of the molecular characteristics of tumor immune interaction. It provides six main analysis modules, allowing users to explore interactively the relationship between immune infiltration and a wide range of factors [Citation24]. We analyzed the expression of RanBP9 in different cancer types, while focusing on the correlation between RanBP9 expression and its copy number changes, the abundance of immune infiltrates in breast cancer, which contained B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils and dendritic cells and tumor purity.
LinkedOmics analysis
The LinkedOMics database (http://www.linkedomics.org/login.php) contains 32 types of cancer from the TCGA project and multiclass data and clinical data of a total of 11,158 patients [Citation25]. The LinkFinder module of LinkedOmics was used to study genes differentially expressed in correlation with RanBP9 in the TCGA BRCA cohort (n = 526).
Pearson’s correlation coefficient was used for statistical analysis of the results, which were presented by the use of volcano plots and heat maps. The Link Interpreter module converts the recognized association into biological context through the pathway and network analysis. Gene Set Enrichment Analysis (GSEA) was used to perform Gene Ontology (GO; celluar component [CC], biological processes [BP] and molecular function [MF]), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, miRNA target enrichment and transcription factor target enrichment. The grade standard is a false discovery rate (FDR) <0.05, and 500 simulations were carried out.
c-BioPortal analysis
The cBio Cancer Genomics Portal (http://cbioportal.org) is a web platform for exploring, visualizing and analyzing multidimensional cancer genomics data [Citation26]. It was used to analyze the alterations of RanBP9 in the invasive breast carcinoma (TCGA, provisional) dataset. The search parameters include mutation, copy number variation (CNV) and the effects of RanBP9 mutation on other common mutated genes. The tab OncoPrint displays an overview of genetic alterations per sample in RanBP9.
GeneMANIA analysis
GeneMANIA (http://www.genemania.org) is a flexible, user-friendly web interface for constructing protein–protein interaction (PPI) network, generating hypotheses about gene function, analyzing gene lists and prioritizing genes for functional assays [Citation27]. GeneMANIA was used to analyze the PPI network of RanBP9 and CTLH complex.
Human tissue microarray
Human tissue arrays (HBreD140Su01) containing 140 breast cancer tissues and 77 adjacent nontumor tissues was purchased from Shanghai Outdo Biotech Company. The experiments were approved by the Independent Ethics Committee of Shanghai Ninth People’s Hospital affiliated to Shanghai JiaoTong University, School of Medicine. The tissue sections were stained with antibody against RanBP9 (1:200, A19238, ABclonal). After staining, ten cases of the 140 breast cancer tissues were found not to meet the requirements and were therefore excluded in the follow-up analysis. The data based on the average immunohistochemistry (IHC) scores (H-scores) were provided by two independent researchers, and statistical analysis was performed by using unpaired t-test. Briefly, H-scores equal the score of density multiply the score of range. A H-score of 0–3 is considered as low expression, and a H-score of 4–12 is considered as high expression. Density score was was determined according to the following criteria: 0 = negative expression, 1 = weak (light brown), 2 = moderate (yellow brown) and 3 = strong (brown). For the score of range: 0 = < 1% expression, 1 = 1–25%, 2 = 25–50%, 3 = 50–75% and 4 = 75–100%.
Results
The expression of RanBP9 in breast cancer
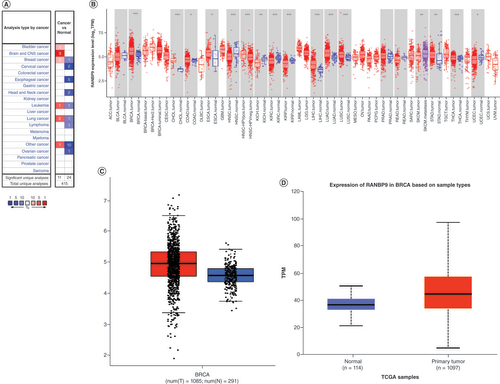
First, we compared the transcriptional levels of RanBP9 mRNA in cancer tissues and their adjacent normal tissues by examining the Oncomine databases (). The expression of RanBP9 is upregulated in the cancer tissues of the brain and the CNS, breast, lung and bladder cancers, while downregulated in esophageal, head and neck, cervical and ovarian cancers (p < 0.05; ). Similar results were also observed from the TIMER database (). Interestingly, the expression of RanBP9 is up-regulated in all the subtypes of breast cancer (luminal, HER-2 overexpression, and basal-like; ). To confirm these results, we further studied the expression of RanBP9 in breast cancer using other datasets. The results from the GEPIA and UALCAN databases revealed that the mRNA level of RanBP9 was much higher in breast cancer than that in normal breast tissues ( & D).
(A) The levels of RanBP9 mRNA in different types of cancer (Oncomine). (B) The levels of RanBP9 mRNA in different types of cancer (TIMER). (C) The expression of RanBP9 in breast cancer (GEPIA). (D) The expression of RanBP9 in breast cancer (UALCAN).
*p < 0.05; **p < 0.01; ***p < 0.001.
TCGA: The Cancer Genome Atlas; TPM: Transcript per million
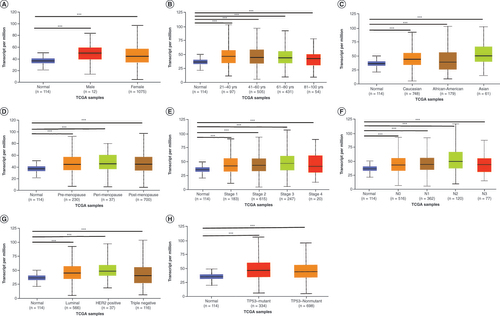
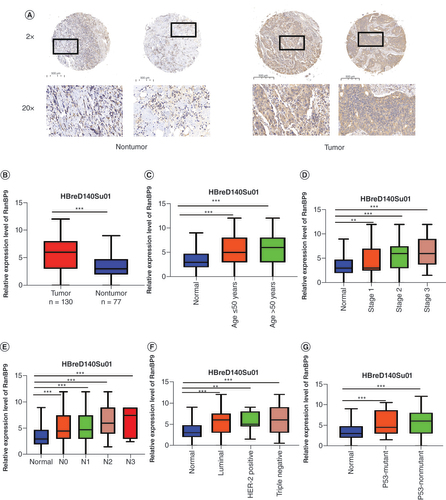
Next, we used the clinicopathological properties to stratify breast cancer patients into various subgroups (). Subgroup analyses of gender, age, race, menopause, tumor stage, lymph node metastasis, subclasses and TP53 mutation status showed that the expression of RanBP9 in breast cancer patients was significantly higher than that in healthy people. The subsequent analysis showed that younger age (), higher stage () and worse status of lymph node metastasis () correlated with higher levels of RanBP9. However, the level of RanBP9 at stage 3 or N4 of breast cancer is slightly lower than that of RanBP9 at stage 2 or N3, and this may be due to the advanced stage of breast cancer and associated factors ( & F).
(A) Boxplot showing relative expression of RanBP9 in normal individuals of either gender. (B) Boxplot showing relative expression of RanBP9 in normal individuals of any age. (C) Boxplot showing relative expression of RanBP9 in normal individuals of any race. (D) Boxplot showing relative expression of RanBP9 in normal individuals based on menopause status. (E) Boxplot showing relative expression of RanBP9 in normal individuals of any cancer stages. (F) Boxplot showing relative expression of RanBP9 in normal individuals based on nodal metastasis. (G) Boxplot showing relative expression of RanBP9 in normal individuals of any subclass. (H) Boxplot showing relative expression of RanBP9 in normal individuals based on TP53 mutation status. Data are mean ± standard error.
*p < 0.05; **p < 0.01; ***p < 0.001.
TCGA: The Cancer Genome Atlas.
The prognostic value of RanBP9 in breast cancer patients
We used Kaplan–Meier Plotter tools and log-rank test analysis to study the relationship between RanBP9 mRNA level and the prognosis of breast cancer patients, leading us to discover that in the patient group with high RanBP9 expression, the RFS, OS, PPS and DMFS were shorter than in those with low expression (HR = 1.4, log rank p < 0.001, HR = 1.4, log rank p = 0.021, HR = 1.97, logrank p < 0.001 and HR = 1.24, logrank p = 0.0063, respectively, –D). These results suggest that RanBP9 overexpression in breast cancer may be associated with poor prognosis.
(A–D) RFS, OS, PPS and DMFS of breast cancer patients grouped by RanBP9 expression level (auto selected the best cutoff value). (E–H) RFS of patients with luminal A, luminal B, HER-2 overexpression, basal-like breast cancer grouped by RanBP9 expression level (auto selected the best cutoff value).
DMFS: Distant metastasis-free survival; HR: Hazard ratio; OS: Overall survival; PPS: Post-progression survival; RFS: Relapse-free survival.
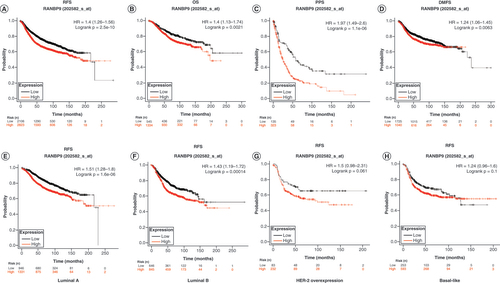
We further explored the relationship between the subtypes of breast cancer and the prognostic values and found that in both luminal A subtype and luminal B subtype, the RFS of patients with high RanBP9 expression was significantly shorter than that of the patients with low expression (HR = 1.51, log rank p < 0.001 and HR = 1.43, log rank p < 0.001, respectively, –F). However, in the HER-2 overexpression subtype or basal-like subtype, there was no significant difference in RFS between the patients with high and low RanBP9 expression (HR = 1.5, log rank p = 0.06 and HR = 1.24, log rank p = 0.1, respectively, & H).
IHC analysis of RanBP9 expression in breast cancer tissues
We next examined the expression of RanBP9 protein in breast cancer tissues to verify the findings from the breast cancer databases. We used IHC to detect RanBP9 protein in a breast cancer tissue array, the clinical characteristics of which are shown in . Consistent with the results from the database analyses, the level of RanBP9 protein in breast cancer tissues was significantly higher than that in adjacent normal breast tissues (). As shown in , the H-score for RanBP9 expression in breast cancer tissues increased significantly compared with normal tissues (p < 0.001). Similar to our earlier analysis of the breast cancer databases (), we also stratified the patients into subgroups based on age, stage, lymph node metastasis, subclasses and P53 mutation status (–G). The results from IHC were consistent with those from the databases (); in these subgroups, the expression of RanBP9 in breast cancer tissues was significantly higher than that in normal tissues. Furthermore, the level of RanBP9 in breast cancer tissues at stages 2 and 3 was higher than that at stage 1 (). Also, the level of RanBP9 gradually increased with the grade of lymph node metastasis, reaching the highest at N3 ().
Table 1. Characteristics of patients with breast cancer included in immunohistochemical analysis.
(A) Representative images for RanBP9 immunohistochemistry using serial sections of tissue microarrays from nontumor and tumor tissues. Magnification: ×2 and ×20 for the insets. (B) A statistical graph showing the score of the results of tissue microarray staining. (C) Boxplot showing relative expression of RanBP9 in normal individuals of any age. (D) Boxplot showing relative expression of RanBP9 in normal individuals of any cancer stages. (E) Boxplot showing relative expression of RanBP9 in normal individuals based on nodal metastasis. (F) Boxplot showing relative expression of RanBP9 in normal individuals of any subclass. (G) Boxplot showing relative expression of RanBP9 in normal individuals based on P53 mutation status. Data are mean ± standard error.
*p < 0.05; **p < 0.01; ***p < 0.001.
N: Node.
After verifying the high-level expression of RanBP9 in breast cancer tissues, we further explored whether the high level is associated with poor prognosis. However, the difference between the two groups was not statistically significant, which may be due to the small sample size or other undefined factors (Supplementary Figure 1).
Genetic & epigenetic changes of RanBP9 in breast cancer
We next used the c-BioPortal online tool to analyze potential genetic and epigenetic alterations of RanBP9 in breast cancer cells. We found that RanBP9 gene was altered in 132 of 1101 patients (12%), including amplification, deep deletion, high mRNA expression, low mRNA expression and missense mutations (). Among the alterations, the most common type was high mRNA expression (). Next, we analyzed the relationship between the types of the alterations and the levels of RanBP9 mRNA. As shown in , among the alterations, the level of RanBP9 mRNA increased from the deletion group, diploid group, gain group to the amplification group in a sequential manner. Thus, RanBP9 amplification causes highest overexpression of RanBP9 ().
(A) RanBP9 gene expression and mutation analysis in breast cancer (c-BioPortal). (B)RanBP9 expression in different RanBP9 copy number variation groups (c-BioPortal). (C) Promoter methylation of RanBP9 in breast cancer (UALCAN).
*p < 0.05.
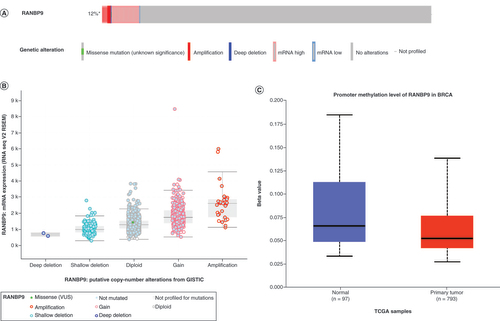
Epigenetic modifications also play an important role in the occurrence and development of cancer. In breast cancer, DNA methylation is linked to the clinicopathological characteristics, such as tumor staging and classification, and it can also affect the progression and prognosis of breast cancer patients [Citation28,Citation29]. We then examined the methylation level of RanBP9 promoter in breast cancer cells. Our analysis of the data in the UALCAN database showed that the methylation level of RanBP9 promoter in breast cancer cells was lower than in normal cells (). The reduction in methylation may cause the levels of RanBP9 to elevate in cancer tissues () and lead to potential poor prognosis ().
Genes co-expressed with RanBP9 & the functional networks in BRCA
To unveil the functions and mechanism of RanBP9 in breast cancer, we first used the Function module of LinkedOmics to analyze the mRNA sequencing data from 526 BRCA patients. Among the genes correlating with RanBP9 expression, 3280 (dark red dots) including NUP153 correlated positively with RanBP9, whereas 2868 (dark green dots) including RPL34 correlated negatively with RanBP9 (FDR <0.01; ), indicating the extensive impact of RanBP9 on the transcriptome. The top 50 significant gene sets positively or negatively correlating with RanBP9 are shown in the heat maps ( & C). Among the genes positively correlating with RanBP9 expression, the top three genes are NUP153 (positive rank no. 1 Pearson’s correlation = 0.5484, p = 1.228e-42), TAF11 (Pearson’s correlation = 0.4539, p = 4.309e-28), and RACGAP1A (Pearson’s correlation = 0.4295, p = 5.032e-25; Supplementary Figure 2A–C).
(A) The genes strongly correlating with RanBP9 as identified by Pearson test in breast cancer. (B & C) Heat maps showing genes positively and negatively correlating with RanBP9 in BRCA (TOP 50). (D–G) Significantly enriched Gene Ontology annotations and Kyoto Encyclopedia of Genes and Genomes pathways of RanBP9 in BRCA.
CC: Cellular component; BP: Biological process; MF: Molecular function.
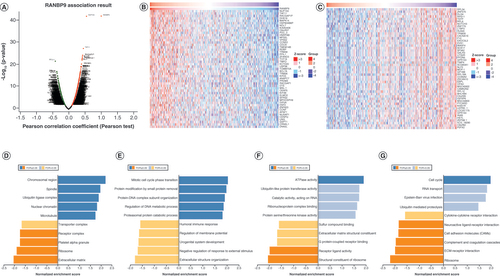
We next utilized GO term annotation by GSEA to analyze the classes of genes differentially expressed in correlation with RanBP9. We found that the genes were primarily associated with chromosomal region, spindle, ubiquitin ligase complex, nuclear chromatin and microtubules ( & Supplementary Table 1), where they are mainly involved in biological processes such as protein modification by mitotic cycle phase transition, small protein removal and protein–DNA complex subunit organization ( & Supplementary Table 2). As shown in & Supplementary Table 3, the genes can promote ATPase activity while inhibiting receptor ligand activity and structural constituent of ribosome in the molecular function MF category. Furthermore, the KEGG pathway analysis unveiled the enrichment in the cell cycle pathways ( & Supplementary Table 4). We also used GSEA to study the potential corresponding miRNAs and transcription factor networks of RanBP9. However, no significant enrichment of miRNAs co-expressed with RanBP9 was found (Supplementary Figure 2D), whereas the enrichment of transcription factors includes V$E2F_03 and RCGCANGCGY V$NRF1_Q6 (Figure S2E).
RanBP9 is linked to the tumor purity & immune infiltration in BRCA
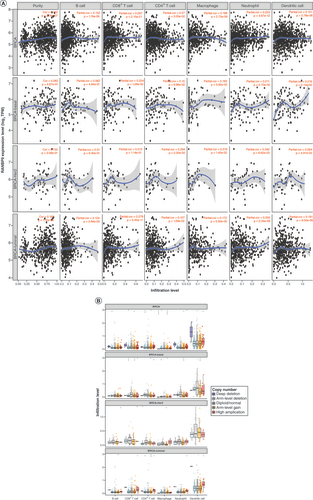
The occurrence and development of tumors are inseparable from the immune system, and the rise of immunotherapy is changing the paradigm of breast cancer treatment. Therefore, to examining the possibility of that RanBP9 might be explored as an immune target in breast cancer, we took advantage of the TIMER database to investigate whether the expression of RanBP9 correlated with the immune infiltration levels. As shown in , the expression of RanBP9 was positively associated with tumor purity (correlation [Cor] = 0.162, p = 2.74e-07). Further analysis of different subtypes of BRCA showed that the expression of RanBP9 correlated with the BRCA tumor purity of the luminal type (Cor = 0.165, p = 1.12e-04), HER-2 overexpression type (Cor = 0.135, p = 3.08e-01) and basal type (Cor = 0.062, p = 4.87e-01; ). We then investigated the correlation between RanBP9 and the infiltration of immune cell types. We found that there was a positive correlation between RanBP9 expression and the infiltration of B cells (Cor = 0.152, p = 1.74e-06), CD8+ T cells (Cor = 0.297, p = 2.15e-21), CD4+ T cells (Cor = 0.07, p = 3.05e-02), macrophages (Cor = 0.188, p = 2.75e-09), neutrophils (Cor = 0.223, p = 3.67e-12) and dendritic cells (Cor = 0.151, p = 2.78e-06) in breast cancer (). Furthermore, the CNV of RanBP9 exhibited a correlation with infiltrating levels of CD8+ T cells, CD4+ T cells, macrophages and neutrophils in breast cancer (). Among different BRCA subtypes, RanBP9 CNV is linked to the infiltrating level of different types of immune cells: B cells and CD4+ T cells in the luminal type, CD8+ T cells and CD4+ T cells in HER2 overexpression type, and T cells, macrophages, neutrophils and dendritic cells in the basal type (). Thus, RanBP9 may affect the immunity of different subtypes of breast cancer and do so through potential distinct mechanisms.
(A) The correlation between the abundance of immune cells and the expression of RanBP9 in BRCA and its subtype. (B)RanBP9 copy number variation affects the infiltrating levels of immune cells in BRCA and its subtype.
*p < 0.05; **p < 0.01; ***p < 0.001.
TPM: Transcript per million.
RanBP9 protein–protein interactions
Finally, we used GeneMANIA to conduct protein–protein interaction (PPI) network analysis to identify the potential interacted proteins with RanBP9. We found that there were 21 nodes and 250 edges in the PPI network, while RanBP9 potentially interacted with 20 proteins including L1CAM, RANBP10, GID8, MAEA, FMR1, NGFR, YPEL5, GRM8, HMBS, OPRM1, S100A7, ENTPD1, HGF, MKLN1, MET, HIPK2, RMND5B, RMND5A, WDR26 and GRM2 (). Interestingly, most members of the CTLH complex, such as GID8, MAEA, MKLN1, RMND5A, RMND5B, RanBP10, WDR26 and YPEL5, are included in the 20 proteins (), consistent with the notion that RanBP9 is part of the CTLH protein complex. We therefore assessed the PPI network of the CTLH complex, leading us to discover proteins, including THSD7B, ENTOD1 and OFD1, that interacted with the complex (). Among them, OFD1, NOVA1, YPEL2, YEPL3, HTRA2, UCK2, PTGER3 and S100A7 are reportedly linked to breast cancer [Citation30–38]. These results set the stage for future studies of the potential functional interaction between RanBP9 and the CTLH complex in breast cancer progression.
Discussion
RanBP9 is involved in the regulation of multiple pathways and cellular processes. Previous studies have shown that RanBP9 inhibits the motility and invasiveness of breast cancer cell lines [Citation39,Citation40]. However, there are limited studies on the mechanisms as to how RanBP9 might modulate the development and prognosis of breast cancer. To gain more detailed insights into the potential functions of RanBP9 in breast cancer and the underlying regulatory network, we performed bioinformatics analysis of published sequencing data to mainly focus on RanBP9 expression, genetic alterations, functional networks, protein interactions and its link to prognosis and immune cell infiltration in breast cancer. We found that the expression of RanBP9 might be related to the prognosis and immune infiltration of breast cancer, indicating that RanBP9 may serve as a marker and potential therapeutic target of breast cancer, laying the foundation for future functional studies of RanBP9.
RanBP9 is highly expressed in breast cancer tissues, and the high-level expression is not limited by factors such as gender, age and menopause. In addition, the expression of RanBP9 in each subtype of breast cancer is significantly higher than that of normal tissues, with the highest increase in the HER-2 subtype. Future experiments should be performed to explore and verify the link between RanBP9 and breast cancer. In addition, the high RanBP9 expression is associated with poor prognosis, suggesting its potential value as a diagnostic and prognostic marker. However, further studies are needed to verify RanBP9 as a diagnostic and prognostic marker in different subtypes of breast cancer.
Cancer is caused by genetic changes and epigenetic modifications, which lead to the transcriptional imbalance of cancer cells and subsequent increase of the proliferation and metastasis [Citation41]. CNV is an important source of genetic variation and a hallmark in the cancer genome [Citation42]. In this study, we found that the major types of RanBP9 alterations are high mRNA and RanBP9 amplification, causing high levels of RanBP9. Because RanBP9 is involved in a variety of biological processes, its alterations may ultimately affect the occurrence and development of breast cancer through undefined molecular mechanisms.
DNA methylation is one of the most important epigenetic modifications and affects the cellular pathways, including apoptosis, cell adhesion, DNA repair and cell cycle regulation. Earlier studies have shown that DNA methylation can guide the treatment and prognosis of breast cancer patients [Citation43,Citation44]. We found that the methylation level of the RanBP9 promoter in breast cancer is lower than that in normal tissues. Whether this is related to the prognosis needs further investigation. Moreover, the reduced methylation of RanBP9 in breast cancer may activate its transcriptional activity and lead to its overexpression.
In this study, we also explored the network of genes related to RanBP9. GO analysis shows they are primarily involved in mitotic cycle phase transition, small protein removal and protein–DNA complex subunit organization. Our results from KEGG analysis further reveal the enrichment of the genes in cell cycle pathways. Hanahan et al. described ten hallmarks of tumors, the first of which is ‘continuous proliferation’ [Citation45]. The role of cell cycle signals in the occurrence and development of breast cancer has been the target of extensive investigation, and related targets and drugs have been developed [Citation46–48]. Therefore, RanBP9 may influence the occurrence and development of tumors by regulating cell cycle activities. Indeed, our enrichmentanalysis of target gene sets using GSEA reveal potential important transcription factors including V$E2F_03 and RCGCANGCGY V$NRF1_Q6, and the function of these molecules is worthy of further investigation.
Immunotherapy has become a new treatment paradigm for tumor therapy. With the continuous exploration of tumor immunotherapy, a variety of targets have been discovered. In addition to the well-known T cells that play a critical role in tumor immunity, other immune cells, such as natural killer cells and neutrophils, also have functions in tumor immunity. Although immunotherapy has achieved significant results in the treatment of many types of tumors, there are still a large number of patients yet to benefit from it, and it is essential to explore new targets and mechanisms to realize the potential. By using tumor purity analysis, we found that RanBP9 is involved in the tumor purity and tumor immunity, while RanBP9 CNV may have correlations with infiltrating levels of various types of immune cells. Our studies establish a link between RanBP9 and tumor immunity. Further studies are clearly needed to elucidate whether RanBP9 can be applied as a useful factor in mediating immunotherapy.
The results from PPI analysis suggest that at the protein level, RanBP9 functions as part of the CTLH complex. The CTLH complex is overexpressed in most of the common cancer types and is involved in many fundamental biological processes, including proliferation, survival, programmed cell death, cell adhesion and migration [Citation9]. Recent findings strongly support the notion that CTLH complex might serve as a functional unit to promote tumor progression and may be key to the understanding of some fundamental biological and pathological processes. RanBP9 is an indispensable component of the CTLH complex, and as such, a deeper understanding of RanBP9 function also requires more studies on the CTLH complex in the future.
It should be noted that our research is mainly based on bioinformatics. Although we confirm the high expression of RanBP9 in breast cancer tissues using tissue microarray analysis, the difference in prognostic analysis is not significant, which may be due to the small sample size or other undefined factors. Therefore, more breast cancer patients are required to verify that the high expression of RanBP9 is associated with poor prognosis. Further, our studies suggest a possible link between RanBP9 and tumor immune infiltration, which should be verified clinically in the future using multiplex IHC and immune cell antigens, such as CD3, CD4, CD8 and Granzyme B. The results from data mining provide the directions for further research, which needs to be conducted using clinical samples in the future to draw more concrete conclusions.
Conclusion
In conclusion, based on data mining and tissue microarray analysis, our research provides multilevel evidence for the relationship between RanBP9 and breast cancer. Our results suggest that RanBP9 is highly expressed in breast cancer tissues and associated with poor prognosis, suggesting its potential value as a diagnostic and prognostic marker. Moreover, RanBP9 exhibit genetic alterations and a decreased methylation level in cancer tissues. We also discovered that RanBP9 might regulate cell cycle progression and is linked to tumor purity and the infiltrating levels of immune cells. These findings lay the foundation for future studies on the potential role of RanBP9 in breast cancer progression.
Future perspective
RanBP9 has a dual effect on tumor progression and is likely involved in the regulation of multiple pathways and cellular processes. Our results pave the way for further research of the relationship between RanBP9 and breast cancer. Future studies should be focused on both the clinical verification and molecular mechanisms. Clinically, more samples need to be collected to analyze how expression of RanBP9 is related to the prognosis and immune infiltration in breast cancer. The mechanism exploration could pay close attention to genetic alterations, functional networks and protein interactions. The results of PPI network indicate that the further study of the role of RanBP9 should be performed in association with the role of the CTLH complex. Together, a detailed delineation of the mechanism by which RanBP9 modulates breast cancer development awaits future experimentation.
RanBP9 has a dual effect on tumor progression. However, the level of RanBP9 transcripts in most tumor samples is higher than in corresponding nontumor samples.
RanBP9 is highly expressed in breast cancer tissues, and the high-level expression is not affected by factors such as gender, age and menopause.
RanBP9 overexpression in breast cancer is associated with poor prognosis, suggesting its potential role as a diagnostic and prognostic marker.
RanBP9 expression is altered in ~12% of patients with invasive breast cancer, and the major types of RanBP9 alterations are high mRNA and RanBP9 amplification, causing high levels of RanBP9.
The reduction in methylation may be the cause of RanBP9 overexpression in cancer tissues and lead to potential poor prognosis.
There are 3280 genes correlating positively with RanBP9, and 2868 correlate negatively with RanBP9, indicating the extensive impact of RanBP9 on the transcriptome.
Functional network analysis suggests that RanBP9 regulates cell cycle.
RanBP9 is involved in the tumor purity and tumor immunity, whereas RanBP9 copy number variation has correlations with infiltrating levels of various types of immune cells.
The results from protein–protein interaction analysis suggest that at the protein level, RanBP9 functions as part of the carboxy-terminal to LisH complex.
Author contributions
Study design and concept: D Li and YL Meng. Data acquisition: YL Meng, YX Ying and MC Zhang. Data analysis and interpretation: YL Meng and YX Ying. Manuscript preparation: YL Meng, YX Ying, MC Zhang, SN Zhang, D Li and Y Yao. Manuscript review: YL Meng, D Li and Y Yao. All authors read and approved the final manuscript.
Ethics conduct of research
The authors state that they have obtained appropriate institutional review board approval by Ethics Committee of Shanghai Ninth People’s Hospital affiliated to Shanghai JiaoTong University, School of Medicine for all human or animal experimental investigations (approval no. YBM-05-02).
Supplemental Document 1
Download MS Word (5 MB)Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.tandfonline.com/doi/suppl/10.2217/epi-2021-0464
Financial & competing interests disclosure
This work was supported by the National Natural Science Foundation of China under grant 81970094; The Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning under TP2015022; Shanghai Pujiang Program under Grant 15PJ1404800. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Additional information
Funding
References
- Sung H , FerlayJ , SiegelRLet al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer Cancer J. Clin.71(3), 209–249 (2021).
- Britt KL , CuzickJ , PhillipsKA. Key steps for effective breast cancer prevention. Nat. Rev. Cancer20(8), 417–436 (2020).
- Zhang B , LiY , LiLet al. Association study of susceptibility loci with specific breast cancer subtypes in Chinese women. Breast Cancer Res. Treat.146(3), 503–514 (2014).
- Perou CM , SørlieT , EisenMBet al. Molecular portraits of human breast tumours. Nature406(6797), 747–752 (2000).
- Goldhirsch A , WoodWC , CoatesASet al. Strategies for subtypes – dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann. Oncol.22(8), 1736–1747 (2011).
- Burstein HJ , SomerfieldMR , BartonDLet al. Endocrine treatment and targeted therapy for hormone receptor-positive, human epidermal growth factor receptor 2-negative metastatic breast cancer: ASCO Guideline Update. J. Clin. Oncol. doi:10.1200/jco.21.01392Jco2101392 (2021) ( Epub ahead of print).
- Moy B , RumbleRB , ComeSEet al. Chemotherapy and Targeted therapy for patients with human epidermal growth factor receptor 2-negative metastatic breast cancer that is either endocrine-pretreated or hormone receptor-negative: ASCO guideline update. J. Clin. Oncol. doi:10.1200/jco.21.01374Jco2101374 (2021) ( Epub ahead of print).
- Rao MA , ChengH , QuayleANet al. RanBPM, a nuclear protein that interacts with and regulates transcriptional activity of androgen receptor and glucocorticoid receptor. J. Biol. Chem.277(50), 48020–48027 (2002).
- Huffman N , PalmieriD , CoppolaV. The CTLH complex in cancer cell plasticity. J. Oncol.2019, 4216750 (2019).
- Maitland MER , OneaG , ChiassonCAet al. The mammalian CTLH complex is an E3 ubiquitin ligase that targets its subunit muskelin for degradation. Sci. Rep.9(1), 9864 (2019).
- Salemi LM , MaitlandMER , MctavishCJ , Schild-PoulterC. Cell signalling pathway regulation by RanBPM: molecular insights and disease implications. Open Biol.7(6), (2017).
- Liu T , RohSE , WooJAet al. Cooperative role of RanBP9 and P73 in mitochondria-mediated apoptosis. Cell Death Dis.4(1), e476 (2013).
- Palmieri D , TessariA , CoppolaV. Scorpins in the DNA damage response. Int. J. Mol. Sci.19(6), (2018).
- Zhao Z , ChengS , ZabkiewiczCet al. Reduced expression of RanBPM is associated with poorer survival from lung cancer and increased proliferation and invasion of lung cancer cells in vitro. Anticancer Res.37(8), 4389–4397 (2017).
- Shao S , SunPH , SatherleyLKet al. Reduced RanBPM expression is associated with distant metastasis in gastric cancer and chemoresistance. Anticancer Res.36(3), 1295–1303 (2016).
- Qin C , ZhangQ , WuG. RANBP9 suppresses tumor proliferation in colorectal cancer. Oncol. Lett.17(5), 4409–4416 (2019).
- Wang D , LiZ , MessingEM , WuG. Activation of Ras/Erk pathway by a novel MET-interacting protein RanBPM. J. Biol. Chem.277(39), 36216–36222 (2002).
- Zou Y , LimS , LeeKet al. Serine/threonine kinase Mirk/Dyrk1B is an inhibitor of epithelial cell migration and is negatively regulated by the Met adaptor Ran-binding protein M. J. Biol. Chem.278(49), 49573–49581 (2003).
- Forbes SA , BeareD , GunasekaranPet al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res.43, D805–811 (2015).
- Tessari A , ParbhooK , PawlikowskiMet al. RANBP9 affects cancer cells response to genotoxic stress and its over expression is associated with worse response to platinum in NSCLC patients. Oncogene37(50), 6463–6476 (2018).
- Rhodes DR , Kalyana-SundaramS , MahavisnoVet al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia9(2), 166–180 (2007).
- Chandrashekar DS , BashelB , BalasubramanyaSaHet al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia19(8), 649–658 (2017).
- Tang Z , LiC , KangBet al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res.45(W1), W98–w102 (2017).
- Li T , FanJ , WangBet al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res.77(21), e108–e110 (2017).
- Vasaikar SV , StraubP , WangJ , ZhangB. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res.46(D1), D956–d963 (2018).
- Gao J , AksoyBA , DogrusozUet al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal.6(269), pl1 (2013).
- Franz M , RodriguezH , LopesCet al. GeneMANIA update 2018. Nucleic Acids Res.46(W1), W60–w64 (2018).
- Fleischer T , TekpliX , MathelierAet al. DNA methylation at enhancers identifies distinct breast cancer lineages. Nat. Commun.8(1), 1379 (2017).
- Jovanovic J , RønnebergJA , TostJ , KristensenV. The epigenetics of breast cancer. Mol. Oncol.4(3), 242–254 (2010).
- Tang Z , ZhuM , ZhongQ. Self-eating to remove cilia roadblock. Autophagy10(2), 379–381 (2014).
- Park HM , KimH , LeeKH , ChoJY. Analysis of opposing histone modifications H3K4me3 and H3K27me3 reveals candidate diagnostic biomarkers for TNBC and gene set prediction combination. BMB Rep.53(5), 266–271 (2020).
- Tang S , ZhaoY , HeXet al. Identification of NOVA family proteins as novel β-catenin RNA-binding proteins that promote epithelial-mesenchymal transition. RNA Biol.17(6), 881–891 (2020).
- Kelemen LE , WangX , FredericksenZSet al. Genetic variation in the chromosome 17q23 amplicon and breast cancer risk. Cancer Epidemiol. Biomarkers Prev.18(6), 1864–1868 (2009).
- Tuttle R , MillerKR , MaioranoJNet al. Novel senescence associated gene, YPEL3, is repressed by estrogen in ER+ mammary tumor cells and required for tamoxifen-induced cellular senescence. Int. J. Cancer130(10), 2291–2299 (2012).
- Wittmann S , BaliP , DonapatySet al. Flavopiridol down-regulates antiapoptotic proteins and sensitizes human breast cancer cells to epothilone B-induced apoptosis. Cancer Res.63(1), 93–99 (2003).
- Shen G , HeP , MaoYet al. Over expression of uridine-cytidine kinase 2 correlates with breast cancer progression and poor prognosis. J. Breast Cancer20(2), 132–141 (2017).
- Zati Zehni A , JeschkeU , HesterAet al. EP3 is an independent prognostic marker only for unifocal breast cancer cases. Int. J. Mol. Sci.21(12), (2020).
- Goh JY , FengM , WangWet al. Chromosome 1q21.3 amplification is a trackable biomarker and actionable target for breast cancer recurrence. Nat. Med.23(11), 1319–1330 (2017).
- Wei JD , KimJY , KimAKet al. RanBPM protein acts as a negative regulator of BLT2 receptor to attenuate BLT2-mediated cell motility. J. Biol. Chem.288(37), 26753–26763 (2013).
- Wei JD , JangJH , KimJH. RanBPM inhibits BLT2-mediated IL-8 production and invasiveness in aggressive breast cancer cells. Biochem. Biophys. Res. Commun.483(1), 305–311 (2017).
- Wu CY , LauBT , KimHSet al. Integrative single-cell analysis of allele-specific copy number alterations and chromatin accessibility in cancer. Nat. Biotechnol.39(10), 1259–1269 (2021).
- Vogelstein B , PapadopoulosN , VelculescuVEet al. Cancer genome landscapes. Science339(6127), 1546–1558 (2013).
- Győrffy B , BottaiG , FleischerTet al. Aberrant DNA methylation impacts gene expression and prognosis in breast cancer subtypes. Int. J. Cancer138(1), 87–97 (2016).
- Absmaier M , NapieralskiR , SchusterTet al. PITX2 DNA-methylation predicts response to anthracycline-based adjuvant chemotherapy in triple-negative breast cancer patients. Int. J. Oncol.52(3), 755–767 (2018).
- Hanahan D , WeinbergRA. Hallmarks of cancer: the next generation. Cell144(5), 646–674 (2011).
- Finn RS , AleshinA , SlamonDJ. Targeting the cyclin-dependent kinases (CDK) 4/6 in estrogen receptor-positive breast cancers. Breast Cancer Res.18(1), 17 (2016).
- De Nonneville A , FinettiP , BirnbaumDet al. WEE1 dependency and pejorative prognostic value in triple-negative breast cancer. Adv. Sci. (Weinh).8(17), e2101030 (2021).
- Freeman-Cook K , HoffmanRL , MillerNet al. Expanding control of the tumor cell cycle with a CDK2/4/6 inhibitor. Cancer Cell39(10), 1404–1421.e1411 (2021).