Abstract
Aim: To perform an epigenome-wide association study (EWAS) of serum folate in maternal blood. Methods: Cross-ancestry (Europeans = 302, South Asians = 161) and ancestry-specific EWAS in the EPIPREG cohort were performed, followed by methyl quantitative trait loci analysis and association with cardiometabolic phenotypes. Replication was attempted using maternal folate intake and blood methylation data from the MoBa study and verified if the findings were significant in a previous EWAS of maternal serum folate in cord blood. Results & conclusion: cg19888088 (cross-ancestry) in EBF3, cg01952260 (Europeans) and cg07077240 (South Asians) in HERC3 were associated with serum folate. cg19888088 and cg01952260 were associated with diastolic blood pressure. cg07077240 was associated with variants in CASC15. The findings were not replicated and were not significant in cord blood.
Tweetable abstract
This research article identified CpG sites associated with serum folate levels in maternal peripheral blood leukocytes. The findings provide new insights about the epigenomic component of serum folate levels.
Folate (vitamin B9) is a coenzyme in the one-carbon metabolism, a pathway that provides one-carbon units for nucleotide biosynthesis and methylation reactions [Citation1,Citation2]. The methyl groups derived from folate are necessary to form S-adenosyl methionine, which is a universal carbon donor for DNA methylation [Citation3]. DNA methylation is an epigenetic mechanism that regulates gene expression and genome stability, as it represses repetitive elements [Citation4]. Adequate folate intake helps to maintain normal DNA methylation levels and minimize DNA damage [Citation5].
It is well recognized that folate is important for tissue growth and fetal development during pregnancy [Citation6]. However, previous studies have suggested that folic acid supplementation is also associated with decreased fasting glucose, fasting insulin and insulin resistance [Citation7], as well as reduced risk for stroke and cardiovascular disease [Citation8]. It has been proposed that DNA methylation could be an important mechanism underlying the observed associations between folate and cardiometabolic traits [Citation9,Citation10]. In line with this hypothesis, a study suggested that low folate levels were associated with hypomethylation in several CpG sites in liver cells of patients with Type 2 diabetes [Citation11].
To date, there is only one epigenome-wide association study (EWAS) of serum folate, which examined the association between maternal serum folate levels and differential DNA methylation in cord blood of their offspring [Citation12]. Another study performed in cord blood evaluated the association between folic acid supplementation and DNA methylation during pregnancy [Citation13]. However, there is no EWAS of serum folate performed in peripheral blood leukocytes.
The epigenetic background of serum folate levels could give valuable insights into folate regulation. Hence, the aims of this study were to perform an EWAS of serum folate in maternal peripheral blood leukocytes, evaluate if the methylation of CpG sites associated with folate were related to cardiometabolic phenotypes, elucidate if methylation of the CpG sites were genetically regulated and evaluate if our findings are replicated in maternal peripheral blood leukocytes with maternal estimated folate intake data assessed with a food-frequency questionnaire (FFQ) as well as in cord blood with maternal serum folate data.
Materials & methods
Study population
The STORK Groruddalen (STORK G) study is a population-based cohort that included 823 healthy pregnant women in the multiethnic area of Groruddalen in Oslo, Norway, from 2008 to 2010. STORK G has been described in detail in previous work [Citation14]. The inclusion criteria were that the woman lived in the study district, planned to give birth at one of the two study hospitals, was less than 20 weeks pregnant, could communicate in Norwegian or any of the eight translated languages and was able to give informed consent. Women were enrolled during early pregnancy, and women with pre-gestational diabetes or in need of intensive hospital follow-up during their pregnancy were excluded. Self-reported ethnic origin was defined by either the individual’s country of birth or their mother’s country of birth if the latter was born outside Europe.
The Epigenetics in Pregnancy (EPIPREG) sample included all women of European (n = 312) or South Asian (n = 168) ancestries participating in STORK G with available DNA [Citation15]. These were the two largest ethnic groups that participated in STORK-G and reflects the ethnic composition of the Groruddalen area of Oslo [Citation15].
STORK G, including the genetic and epigenetic substudies (EPIPREG), was approved by the Norwegian Regional Committee for Medical Health Research Ethics South East (ref. no. 2015/1035). Written informed consent was given by all participants.
Folate measurement & folic acid supplementation
Folate was measured in biobanked serum collected at gestational week 28 with electrochemiluminescence (Roche Diagnostics International, Risch-Rotkreuz, Switzerland) at Medical Biochemistry, Oslo University Hospital. The assay measures the total folate present in serum, and has good affinity for 5-methyltetrahydrofolate (the most common form found in serum), folic acid and other forms of folate [Citation16]. Folate deficiency was determined at < 7 nmol/l [Citation17].
Between gestational weeks 17 and 19 the women were asked whether they used folic acid supplements before pregnancy [Citation18]. Women with missing values were imputed as nonsupplement users. Women were also asked if they used folic acid supplements within 14 days of their appointment at gestational week 28.
Cardiometabolic phenotypes
The data of the cardiometabolic phenotypes used in this study were collected and measured in gestational week 28. Venous blood was drawn into EDTA tubes. The samples were then either biobanked or subject to further preparation and analyses. All women underwent a 75-g oral glucose tolerance test. Gestational diabetes mellitus was diagnosed with the WHO 1999 criteria (fasting glucose ≥7.0 mmol/l and/or 2-h glucose ≥7.8 mmol/l). Fasting insulin was measured with noncompeting immunofluorometric assays (DELFIA, PerkinElmer Life Sciences, Wallac Oy, Turku, Finland). Insulin resistance was estimated by the homeostasis model of insulin resistance, using the HOMA2 calculator version 2.2.2 (www.dtu.ox.ac.uk/homacalculator) based on fasting glucose (HemoCue, Angelholm, Sweden) and C-peptide (DELFIA, PerkinElmer Life Sciences, Wallac Oy) [Citation19]. Fasting plasma total cholesterol, high-density lipoprotein (HDL) cholesterol and triglycerides levels were measured with a colorimetric method (Vitros 5.1 fs, Ortho Clinical Diagnostics, Neckargemünd, Germany) at Akershus University Hospital, Lørenskog, Norway. Low-density lipoprotein cholesterol was calculated with the Friedewald formula [Citation20,Citation21].
Anthropometric data in the present study were also measured at gestational week 28. Systolic and diastolic blood pressure were measured with M6 Comfort HEM-7000-E (Omron, Kyoto, Japan). BMI was calculated from measured height using a fixed stadiometer and body weight (Tanita-BC 418 MA, Tanita Corporation, Tokyo, Japan).
Smoking assessment
Smoking status was evaluated with an interviewer-administered questionnaire and collapsed into two categories for statistical analyses: smokers (current and smokers during the last 3 months before pregnancy) versus nonsmokers (former smokers and never smokers).
DNA methylation profiling & genotyping
DNA was extracted consecutively throughout the data collection, at the Hormone Laboratory, Oslo University Hospital, using a salting out procedure [Citation22]. The genome-wide DNA methylation measurement and genotyping were performed at the Department of Clinical Sciences, Clinical Research Centre, Lund University, Malmö, Sweden, and have been described in detail elsewhere [Citation15].
DNA methylation was quantified in peripheral blood leukocytes using the Infinium MethylationEPIC BeadChip (Illumina, CA, USA). The Meffil R package [Citation23] was used for quality control (QC). 472 individuals from the 480 available and 864,560 probes passed the QC. Blood cell composition (CD8T, CD4T, NK, monocytes, B cells and neutrophils) were calculated with Meffil using the Houseman reference panel [Citation24] during the QC procedure. Lastly, Y and X chromosome probes that harbor SNPs and cross-reactive probes per Pidsley et al.‘s list [Citation25] were not considered for statistical analyses. Thus, the EWAS analyses performed are based on 792,530 probes in total.
Technical validation showed a good agreement between the Infinium MethylationEPIC BeadChip and pyrosequencing on four CpG sites preliminary associated with fasting glucose, 2-h glucose and BMI (n = 30). Details of the technical validation procedure are available in a previous publication [Citation15].
The Illumina CoreExome chip was used for genotyping and PLINK 1.9 software [Citation26] for QC and variant filtering. Variants with deviations of Hardy–Weinberg equilibrium (p = 1.0 × 10-6), low call rate (<95%) and low minor allele frequencies (MAF), were removed. After variant filtering, genetic ancestry origin was assessed through principal component analysis by using the variance-standardized relationship matrix using PLINK 1.9. There was a clear separation between Europe and South Asia, which corresponded to reported ancestry [Citation15]. Approximately 300,000 variants were left for imputation, and 438 women (300 European and 138 South Asian) passed the QC.
Imputation for Europeans and South Asians was done by mapping the genome-wide association study scaffold to NBI build 37 of the human genome. For each ancestry, their correspondent 1000 genomes project panel was used (Phase 3, www.well.ox.ac.uk/~wrayner/tools/) [Citation27], using IMPUTE2 (version 2.3.2) [Citation28]. PLINK 1.9 was used for a postimputation QC. Non-SNP variants and low-quality postimputation SNPs (info <0.9), SNPs with MAF <5% and variants that deviated from Hardy–Weinberg equilibrium (p < 1.0 × 10-6) were removed.
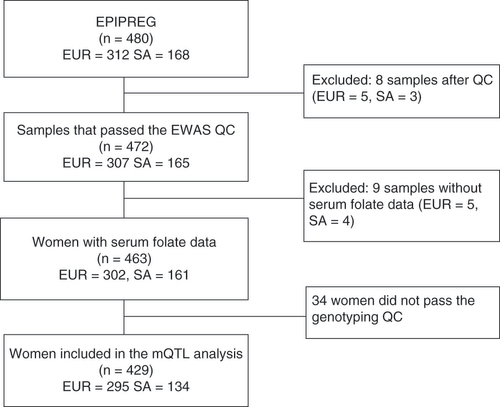
Study flow
For the EWAS studies, 463 women (302 European and 161 South Asians) were included who passed the EWAS QC procedure and had serum folate available (). From these, 295 samples from Europeans and 134 South Asians also passed the genome-wide association study QC and could be included in the methyl quantitative trait loci (mQTL) analysis ().
Statistics
Beta-values were transformed to M-values [Citation29]. For the cross-ancestry EWAS of serum folate levels, linear mixed models using the R packages lme4 and lmerTest [Citation30] were used, calculating the p-values with the Satterthwaite method. The M-values were the outcome, and serum folate the exposure. Age, smoking and blood cell composition were included in the model as fixed effects and ancestry as random intercept to account for potential ancestry-related differences in DNA methylation and folate levels. Ancestry-specific EWAS were performed separately in Europeans and South Asians using standard multivariate linear models using the limma R package [Citation31], adjusting for age, smoking and blood cell composition. For EWAS, a false discovery rate (FDR) threshold of 5% was used.
The residuals of CpG sites that passed the FDR threshold were inspected. If a CpG site had extreme methylation outliers, methylation values below the 2nd and above 98th percentiles were replaced with less extreme values with the winsorization method implemented in the R package ‘broman.’
For the Gene Ontology (GO) analysis, an over-representation test was performed using the nominal significant CpG sites (p-value < 1.0 × 10-4) with the web software WebGestalt [Citation32]. This was done for each EWAS analysis. ‘Biological process non-redundant’ was used as the database of reference, and only GO terms that were composed of at least five genes were included. The 20 most significant GO terms (uncorrected p-value < 0.05) are reported, and those that passed a 5% FDR are noted.
For CpG hits from the cross-ancestry EWAS, their associations with cardiometabolic traits were assessed using linear mixed models and the same covariates as in the discovery analysis. For CpG hits from the ancestry-specific EWAS, associations with cardiometabolic traits were tested using standard linear regression models restricted to the same ancestry, including the same covariates from the corresponding EWAS. A p-value <0.05 was accepted for these analyses.
The GEM R package [Citation33] was used for mQTL analysis. For the cross-ancestry CpG sites, the analysis was performed separately in Europeans and South Asians, adjusting for age, smoking and blood cell composition, followed by a fixed effect meta-analysis. For ancestry-specific CpG sites, the corresponding ancestry was analyzed using standard multivariate models adjusted for the aforementioned covariates. A standard genome-wide p-value threshold of 5 × 10-8 was used. The mQTLs were defined as cis-mQTLs if they were located ± <1 Mb from the methylation site; otherwise, they were classified as trans-mQTLs. The web-tool LD-link (https://ldlink.nci.nih.gov/?tab=home) was used to evaluate whether the mQTLs were in linkage disequilibrium (LD) defined as R2 > 0.8 [Citation34]. Utah residents from North and West Europe (CEU) were used as reference population for Europeans, and the Punjabi from Lahore, Pakistan (PJL) and Sri Lankan Tamil from the UK (STU) for South Asians. If the mQTLs of a CpG site were in high LD within each other, only the most significant variant was tested for association with cardiometabolic phenotypes. If the mQTL correspond to a cross-ancestry CpG site, mixed linear models with ancestry as random intercept for the SNP-phenotype analysis were used; otherwise, the analysis was done in the corresponding ancestry where the CpG was found by using simple linear models.
The R package mediation [Citation35] for mediation analysis was used to assess whether serum folate mediated the relationship between an mQTL and DNA methylation. Cell composition, age and smoking were included as covariates, and 20,000 simulations were used to calculate the confidence intervals using a quasi-Bayesian approximation. A p-value <0.05 threshold was used for these analyses. The mediation analysis calculates the following models: the average causal mediation effects (ACME), which calculate the effect sizes when taking into account the mediator; and the average direct effect (ADE), which represents the effect size when adjusting for the mediator (i.e., removing its effect); and the total effect, which consists of the sum of ACME and ADE effect sizes.
Validation in independent cohorts
Replication of our results was done by using cross-sectional data of estimated folate intake calculated from an FFQ and DNA methylation of maternal peripheral blood leukocytes in two separate subsamples from the Norwegian Mother, Father and Child Cohort Study (MoBa) [Citation36,Citation37]. The first subsample consisted of 1111 mothers and 870 women who underwent assisted reproductive technology (n = 1981). The second subsample included a total of 1022 mothers. Detailed methods and statistics used in MoBa are provided in the Supplementary Material.
Whether these findings had consistent effect sizes and p < 0.05 in an EWAS of maternal serum folate in cord blood from a meta-analysis of a subsample of MoBa (N = 1275) and Generation R (n = 713) cohorts [Citation12] was also evaluated. Details about the data analysis can be found in the source publication [Citation12], but briefly, robust linear regression models were performed, adjusting for maternal age, maternal education, maternal sustained smoking during pregnancy, parity, batch and blood cell composition calculated with the Houseman method. Maternal serum folate data was gathered at gestational week ∼12 in Generation R and ∼18 in MoBa.
It was evaluated whether any of the 443 CpG (FDR < 0.05%) sites reported by Joubert et al. [Citation12] had a consistent direction of the effect compared with EPIPREG. Four hundred ten of the 443 CpG sites were available in both EPIPREG and the cord blood study. A CpG site was considered consistent in both studies if the CpG site had the same direction of the effect across both tissues and passed a Bonferroni p-value threshold (0.05/410). CpG sites that reached a nominal p < 0.05 are reported.
Consultations in public databases
The EWAS catalogue (http://ewascatalog.org/) [Citation38] was consulted to see whether the found CpG sites have been previously associated with folate or any cardiometabolic trait in peripheral blood leukocytes. The catalogue reports associations with a p < 1 × 10-4. mQTLdb (www.mqtldb.org/) [Citation39] was used to identify whether the found CpG sites had mQTLs in maternal peripheral blood leukocytes. The GoDMC database (www.godmc.org.uk/) [Citation40], which consists of a meta-analysis of several cohorts with methylation at peripheral blood leukocytes, was also used to look for mQTLs. mQTLs that passed a standard p < 5 × 10-8 threshold are reported. PhenoScanner (www.phenoscanner.medschl.cam.ac.uk/) [Citation41] was used to identify whether the mQTLs identified in the current sample or public databases were previously associated (p < 5 × 10-8) with folate or any cardiometabolic phenotype.
Results
Population characteristics
Clinical characteristics of the women included are shown in . The mean (standard deviation) age of the participants was 29.9 (4.6) years, and the serum folate level was 14.5 (7.0) nmol/l. Whereas 22.9% reported folate supplement use before pregnancy, only 9.5% reported folate supplements intake at the time of examination in gestational week 28. There were weak correlations between serum folate levels and folate supplement usage before conception (r = 0.10; p = 0.030), folic acid supplementation at week 28 (r = 0.38; p = 2.66 × 10-16) and folate deficiency (r = -0.23; p = 2.21 × 10-7).
Table 1. Clinical characteristics in gestational week 28 of the women included in the study.
Discovery analysis in EPIPREG
In the cross-ancestry EWAS, increased methylation of cg19888088 (annotated to EBF3) and cg10871182 was associated with lower levels of serum folate ( & ). There was no evidence of inflation (λ = 1.05). Summary statistics of cross-ancestry CpG sites with p < 1.0 × 10-4 are presented in Supplementary Table 1.
Each dot represents a CpG with its chromosome position on the x-axis, and the significance level [depicted as -log10(p)] on the y-axis. The dashed line represents p = 1.0 × 10-4 and the solid line a false discovery rate of 5%. The CpG sites cg19888088 and cg10871182 passed the false discovery rate threshold.
![Figure 2. Manhattan plot of the cross-ancestry epigenome-wide association study. Each dot represents a CpG with its chromosome position on the x-axis, and the significance level [depicted as -log10(p)] on the y-axis. The dashed line represents p = 1.0 × 10-4 and the solid line a false discovery rate of 5%. The CpG sites cg19888088 and cg10871182 passed the false discovery rate threshold.](/cms/asset/57eef3b8-ca2d-4603-a3d8-8ff2fc6de366/iepi_a_12324452_f0002.jpg)
Table 2. Summary statistics and genomic location of the CpG sites that passed the false discovery rate <0.05 in each epigenome-wide association study.
In the ancestry-specific EWAS, cg01952260 in Europeans ( & Supplementary Figure 1A) and cg07077240 (annotated in HERC3) in South Asians ( & Supplementary Figure 1B) were associated with reduced methylation at higher serum folate. No evidence of inflation in either the European (λ = 1.07) or South Asian EWAS (λ = 1.06) was found. Summary statistics of CpG sites with p < 1 × 10-4 from the ancestry-specific EWAS are presented in Supplementary Tables 2 3 for Europeans and South Asians, respectively.
Residual inspection of the models (Supplementary Figure 2) suggested that the association with cg10871182 was driven by outliers. After applying winsorization, the direction of the effect of the association persisted but the p-value was attenuated (effect = -0.012, standard error [SE] = 0.0028, p-value = 1.88 × 10-5), thus it was not followed for further analyses.
GO enrichment analysis
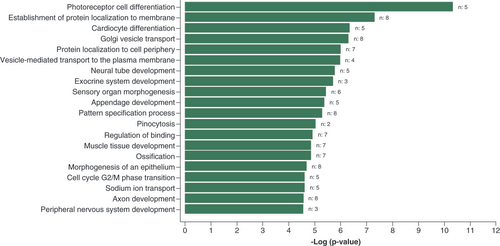
In the GO enrichment analysis of 121 CpG sites with p < 1 × 10-4 from the cross-ancestry EWAS, only the GO-term ‘photoreceptor cell differentiation’ passed the FDR <0.05 threshold ( & Supplementary Table 4). Using the CpG sites with p < 1 × 10-4 from the European EWAS (n = 77) (Supplementary Table 5), no GO term above the FDR threshold was identified. Lastly, performing the same analysis in the South Asian EWAS (n = 141), enrichment (FDR < 0.05) in ‘cell-cell adhesion via plasma-membrane adhesion molecules’ and ‘protein localization to cell periphery’ were identified (Supplementary Table 6).
Associations with cardiometabolic traits
cg19888088 identified in the cross-ancestry EWAS (effect size = -0.002, SE = 0.001, p = 0.0356), and cg01952260 identified in the European EWAS (effect size = 0.005, SE = 0.002, p = 0.022) were associated with diastolic blood pressure in EPIPREG (n = 457). Methylation levels of cg07077240 were not related to any of the other phenotypes tested (Supplementary Table 7).
According to the EWAS catalogue, cg19888088, cg01952260 and cg07077240 have not previously been associated with folate nor any cardiometabolic phenotype.
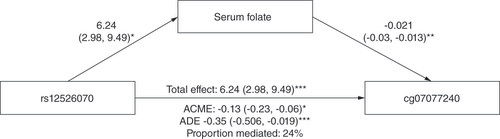
Genetic variants & DNA methylation
In EPIPREG, mQTL analysis identified gene variants only for cg07077240 (rs12526070 and rs113796915) (). Because the variants were in strong LD with each other (R2 = 0.966) only rs12526070 was analyzed further for the phenotype associations. In EPIPREG, the minor allele of rs12526070 (T) was associated with increased serum levels of folate (p = 9.19 × 10-5), triglycerides (p = 0.006), total cholesterol (p = 0.041) and HDL cholesterol (p = 0.023) (Supplementary Table 8). No associations between rs12526070 and serum folate levels nor with any cardiometabolic phenotypes in PhenoScanner were found.
Table 3. Methyl quantitative trait loci associated with methylation at the folate related CpG sites.
In mQTLdb, we found 4 trans-mQTL for cg07077240 (Supplementary Table 9) in LD within each other (R2 from 0.966 to 0.974). The most significant variant was rs4268751 locaed in HS3ST4. The variants rs112030070 and rs112571402 (R2 = 1) located in WDR76 were trans-mQTLs for cg01952260 (Supplementary Table 9). GoDMC has not reported mQTLs for these CpG sites. PhenoScanner has not reported associations with folate nor any cardiometabolic phenotype for rs4268751 and rs112030070.
Mediation analysis
Mediation analysis suggested that serum folate explained 24% of the relationship between rs12526070 and cg07077240 (). However, the effect size and p-value of the ADE (Average direct effects) model were stronger in comparison to the average causal mediation effects (ACME) model ().
Replication with folate intake data from MoBa
Evidence of replication in any of the three CpG tested was not found (Supplementary Table 10).
Shared signatures between cord blood & maternal blood DNA methylation
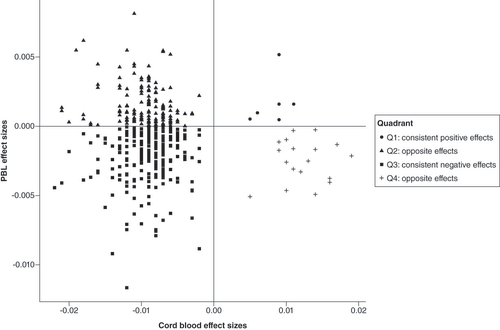
The three CpG sites identified in EPIPREG were not significant in cord blood (Supplementary Table 10). Further, none of the CpG sites associated with serum folate in cord blood reached the Bonferroni threshold in EPIPREG, but 22 CpG sites had the same direction of the effect across both tissues and a nominal p < 0.05 in the cross-ancestry analysis (Supplementary Table 11). In the European and South Asian analyses, three and 16 CpG sites were found that had consistent effects and a nominal p < 0.05, respectively (Supplementary Tables 12 & 13). As shown in , 60% of the 410 CpG sites evaluated had a consistent direction of the effect across peripheral blood leukocytes and cord blood (59% negative and 1% positive effect sizes).
Quadrant 1 (Q1) contains CpG sites with consistent positive effect sizes across both studies (filled circles). Quadrant 2 (Q2) contain CpG sites with negative effect sizes in cord blood, but positive in EPIPREG (filled triangles). Quadrant 3 (Q3) contain CpG sites with consistent negative effect sizes across both studies (filled squares). Quadrant 4 (Q4) contain CpG sites with positive effect sizes in cord blood, but negative effect sizes in EPIPREG (plus symbol).
PBL: Peripheral blood leukocytes.
Discussion
Methylation in maternal peripheral blood leukocytes at cg19888088 (in EBF3), cg01952260 and cg07077240 (in HERC3) were inversely associated with maternal serum folate levels. Methylation at cg19888088 and cg01952260 were associated with diastolic blood pressure. rs12526070, mQTL of cg07077240 annotated to CASC15 was associated with serum folate, triglycerides, total cholesterol and HDL cholesterol. Mediation analysis suggested that folate could mediate the relationship between rs12526070 and methylation levels of cg07077240 but suggested that other unknown additional factors are likely to explain the relationship. Finally, none of the three CpG sites were replicated.
As no cohort had both serum folate and DNA methylation in the same individuals, we attempted replication in two samples with estimated folate intake from an FFQ and DNA methylation in the same women. We did not replicate our findings using folate intake data from MoBa. However, serum folate and estimated folate intake from FFQs are modestly correlated [Citation42–44], as FFQs are hampered by inaccurate reporting and other method limitations [Citation45]. Therefore, the lack of replication could be due to underlying methodological differences across folate measurement methods.
We further evaluated if the CpG sites identified in EPIPREG were also significant (p < 0.05) in an EWAS of maternal serum folate in cord blood of their offspring. The CpG sites were not significant in cord blood DNA methylation. The last could be because maternal folate levels do not entirely reflect offspring folate levels – that is, DNA methylation and serum folate were not measured in the same individual. The transport of maternal folate to cord blood depends on the activity of several transporters in the placenta [Citation46]. During gestation, folate is accumulated in the placenta and cord blood to ensure enough folate to the fetus [Citation46,Citation47], thus folate levels in the fetus can be higher than maternal folate levels. In this context, maternal folate levels are an indirect measurement of folate in cord blood, which may hamper our efforts to find common associations across studies. In addition, the lack association could also be due to differing leukocyte composition between peripheral blood and cord blood [Citation48], and underlying differences between the design of both studies, such as data collected at different time points (gestational week 28 in EPIPREG, <20 week in the cord blood EWAS), and the selection of covariates for the discovery analysis. Lastly, in the quadrant plot presented in , we observed that 60% of the CpG sites identified in cord blood had the same direction of effects in EPIPREG. A better powered study is necessary to verify whether some of these potential shared loci could be formally replicated.
The cross-ancestry site cg19888088, located in the EBF3 gene, has not previously been associated with serum folate levels. However, DNA methylation at two other CpG sites within EBF3 (cg26229752 and cg17726092) in cord blood have been inversely associated with maternal serum folate levels [Citation12]. Thus, we speculate that maternal folate levels are associated with lower EBF3 methylation levels. EBF3 is a transcription factor that influences the cerebral cortex’s laminal formation [Citation49]. Loss of function mutations in EBF3 have been associated with developmental defects such as altered neural development [Citation50], and phenotypes such as intellectual disability, facial dysmorphism, ataxia and autism [Citation49,Citation51]. Evidence further suggests that EBF3 may act as a tumor suppressor [Citation52]. Both aberrant DNA methylation patterns [Citation52,Citation53] and gene silencing in EBF3 [Citation52] are associated with several types of cancers.
The site cg01952260 found in Europeans and cg07077240 found in South Asians have not previously been associated with folate. Decreased DNA methylation at cg01952260 in naive CD4+ T cells has been associated with systemic lupus erythematosus [Citation54]. cg07077240 is located in the HERC3 gene, that negatively regulates the NF-Kb pathway which is an important activator of inflammatory and immune reactions [Citation55]. Folate is a vital nutrient for regulation of the immune reaction and inflammatory response [Citation56]. Therefore, methylation at these CpG sites could potentially play a role in inflammatory related processes associated with folate.
Methylation levels of cg01952260 and cg07077240 were associated with diastolic blood pressure in EPIPREG. Folate has previously been associated with blood pressure [Citation57,Citation58]. Furthermore, a meta-analysis found that intake of folic acid in pregnancy lowered preeclampsia risk – specifically through multivitamins containing folic acid [Citation59] or other folic acid supplements [Citation60]. Nevertheless, only Yadav et al. [Citation61] has suggested that DNA methylation could be an intermediate mechanism between folate metabolism and blood pressure. The study found that global hypomethylation was associated with blood pressure in a North Indian population. The association was more pronounced in nonmedicated hypertensive individuals carrying the T allele of the variant rs1801133 located in MTHFR [Citation61]. Interestingly, another study found that BMI was associated with DNA methylation in the MTHFS gene, an enzyme relevant to folate metabolism [Citation62]. Hence, folate regulation could be associated with DNA methylation patterns that in turn were associated with other cardiometabolic related traits. However, in the EWAS catalogue, we did not find previous associations between blood pressure and methylation at cg01952260 and cg07077240. Hence, these finding should be corroborated in other studies.
rs12526070, mQTL of cg07077240, was associated with several lipid parameters, which have not been reported previously. However, in pregnancy, the lipid profile fluctuates, with a decrease in the first trimester followed by a subsequent increase as pregnancy progresses [Citation63]. Therefore, it remains to be seen whether the associations between rs12526070 and lipids are related to the lipid traits themselves or represent pregnancy-related lipid changes. The mQTLs previously reported for cg07077240 and cg01952260 in mQTLdb were not replicated in our mQTL analysis or in the GoDMC database. Further studies are needed to verify the mQTLs associated with cg07077240 and cg01952260.
The mediation analysis implied that the T allele of rs12526070 increases methylation levels at cg07077240 with serum folate as a mediator. Nevertheless, the ADE model was stronger than the ACME model, implying that it is highly likely that other unknown factors are likely to mediate this relationship aside of folate. Unfortunately, we lack independent variants to perform a robust Mendelian randomization analysis to establish causal relationships.
A major strength of this study is the well characterized cohort, which allowed association analyses with other cardiometabolic phenotypes. Further, genetic data allowed us to perform mQTL analysis to find potential gene variants related to DNA methylation of the CpG sites. An important limitation of this work is the small sample size, which only allowed detection of moderate to strong effect sizes. Finally, we lacked a proper replication cohort with DNA methylation in peripheral blood leukocytes and serum folate data in the same individuals.
Conclusion
We identified three novel CpG sites associated with serum folate levels in peripheral blood leukocytes. The cross-ancestry CpG site cg19888088 was located in the EBF3 gene, a gene related to neural development. cg01952260, found in Europeans, and cg07077240 (in HERC3) found in South Asians, are potentially associated with immune processes related to inflammation. Methylation at cg19888088 and cg01952260 was associated with diastolic blood pressure. Only cg07077240’s was associated with genetic variants, and its mQTL, rs12526070 (in CASC15), was associated with serum folate and lipids. Overall, our findings provide insights about the epigenetic component of serum folate levels.
Three CpG sites associated with serum folate in peripheral blood leukocytes were identified: cg19888088 (cross-ancestry), cg01952260 (in Europeans) and cg07077240 (in South Asians).
cg19888088 is located in the EBF3 gene, which has been implicated in neural development, and methylation at EBF3 has been previously associated with maternal serum folate levels in cord blood.
cg01952260 and cg07077240 in HERC3 are potentially associated with immune processes related to inflammation.
cg19888088 and cg01952260 were associated with diastolic blood pressure in EPIPREG.
cg07077240’s mQTL, rs12526070 (annotated in CASC15), was associated with serum folate and lipids.
A proper replication cohort with both serum folate and DNA methylation data in peripheral blood leukocytes is needed to verify our findings.
Author Contributions
N Fragoso-Bargas and C Sommer contributed to the study conceptualization and design of this substudy. C Sommer and KI Birkeland conceptualized and designed the EPIPREG sample. N Fragoso-Bargas conducted the statistical analyses in EPIPREG, drafted the manuscript, and performed the postimputation quality control. CM Page performed the replication analysis in MoBa. BR Joubert and SJ London provided the summary statistics of cord blood. SL Ødegård curated EPIPREG data. RB Prasad facilitated the wet lab experiments for the methylation and genotyping chips. GH Moen performed the quality control of the genomic data and imputation in EPIPREG. KI Birkeland and AK Jenum designed the STORK-G project. L Sletner contributed with data acquisition in STORK-G. C Sommer is the guarantor of this work, had access to the data and accepts full responsibility for the conduct of the study. All coauthors reviewed/edited the manuscript and approved the final version.
Ethical conduct of research
STORK G, including the genetic and epigenetic substudies (EPIPREG), was approved by the Norwegian Regional Committee for Medical Health Research Ethics South East (ref. no. 2015/1035). Written informed consent was given by all participants.
Supplemental Document
Download MS Word (1.2 MB)Acknowledgments
We thank the women who participated in the STORK Groruddalen study; Maria Sterner, Malin Neptin and Gabriella Gremsperger at the Genomics Diabetes and Endocrinology CRC, Malmö, for the wet lab experiments of the bead chips; and Leif C. Groop, Lund University Diabetes Centre, Malmö, Sweden, for facilitating the wet lab experiments.
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.tandfonline.com/doi/suppl/10.2217/epi-2022-0427
Financial & competing interest disclosure
EPIPREG is supported by the South-Eastern Norway Regional Health Authority (grant number: 2019092), and the Norwegian Diabetes Association. The Norwegian Mother, Father and Child Cohort Study is supported by the Norwegian Ministry of Health and Care Services and the Ministry of Education and Research. The authors are grateful to all the participating families in Norway who take part in this ongoing cohort. The work in MoBA was partly funded by the Norwegian research council’s Centres of Excellence funding scheme, project no. 262700. The work in MoBa was also partially supported by NIH/NIEHS contract no. N01-ES-75558, the Intramural Research Program of the NIH, NIEHS (ZO1 ES49019 to SJ London) and the National Institutes of Health (NIH) Office of Dietary Supplements. GH Moen is the recipient of an Australian Research Council Discovery Early Career Award (project no: DE220101226) funded by the Australian government and supported by the Research Council of Norway (project grant: 325640). SJ London is supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (ZO1 ES49019). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Data sharing statement
Because of strict regulations for genetic data and privacy protection of patients in Norway, all requests for data access are processed by the STORK Groruddalen project’s steering committee. Please contact the principal investigator (PI) of STORK Groruddalen ([email protected]) or the PI of EPIPREG ([email protected]).
Data from the Norwegian Mother, Father and Child Cohort Study and the Medical Birth Registry of Norway used in this study are managed by the national health register holders in Norway (Norwegian Institute of Public Health) and can be made available to researchers, provided approval from the Regional Committees for Medical and Health Research Ethics, compliance with the EU General Data Protection Regulation and approval from the data owners. The consent given by the participants does not open for storage of data on an individual level in repositories or journals. Researchers who want access to data sets for replication should apply through helsedata.no. Access to data sets requires approval from the Regional Committee for Medical and Health Research Ethics in Norway and an agreement with MoBa.
Additional information
Funding
References
- Friso S , UdaliS , DeSantis D , ChoiSW. One-carbon metabolism and epigenetics. Mol. Aspects. Med.54, 28–36 (2017).
- Rosenzweig A , BlenisJ , GomesAP. Beyond the Warburg effect: how do cancer cells regulate one-carbon metabolism?Front. Cell Dev. Biol.6, 90 (2018).
- Irwin RE , PentievaK , CassidyTet al. The interplay between DNA methylation, folate and neurocognitive development. Epigenomics8(6), 863–879 (2016).
- Langie SA , KoppenG , DesaulniersDet al. Causes of genome instability: the effect of low dose chemical exposures in modern society. Carcinogenesis36(Suppl. 1), S61–S88 (2015).
- Krishna SM , DearA , CraigJMet al. The potential role of homocysteine mediated DNA methylation and associated epigenetic changes in abdominal aortic aneurysm formation. Atherosclerosis228(2), 295–305 (2013).
- Virdi S , JadavjiNM. The impact of maternal folates on brain development and function after birth. Metabolites12(9), 876 (2022).
- Zhao JV , SchoolingCM , ZhaoJX. The effects of folate supplementation on glucose metabolism and risk of type 2 diabetes: a systematic review and meta-analysis of randomized controlled trials. Ann. Epidemiol.28(4), 249–257 (2018).
- Li Y , HuangT , ZhengYet al. Folic acid supplementation and the risk of cardiovascular diseases: a meta-analysis of randomized controlled trials. J. Am. Heart. Assoc.5(8), e003768 (2016).
- Crider KS , YangTP , BerryRJ , BaileyLB. Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate’s role. Adv. Nutr.3(1), 21–38 (2012).
- Kok DE , SteegengaWT , McKayJA. Folate and epigenetics: why we should not forget bacterial biosynthesis. Epigenomics10(9), 1147–1150 (2018).
- Nilsson E , MatteA , PerfilyevAet al. Epigenetic alterations in human liver from subjects with type 2 diabetes in parallel with reduced folate levels. J. Clin. Endocr. Metab.100(11), E1491–E1501 (2015).
- Joubert BR , den DekkerHT , FelixJFet al. Maternal plasma folate impacts differential DNA methylation in an epigenome-wide meta-analysis of newborns. Nat. Commun.7, 10577 (2016).
- Irwin RE , ThursbySJ , OndicovaMet al. A randomized controlled trial of folic acid intervention in pregnancy highlights a putative methylation-regulated control element at ZFP57. Clin. Epigenetics11, 31 (2019).
- Jenum AK , SletnerL , VoldnerNet al. The STORK Groruddalen research programme: a population-based cohort study of gestational diabetes, physical activity, and obesity in pregnancy in a multiethnic population. Rationale, methods, study population, and participation rates (vol 38, pg 60, 2010). Scand. J. Public Health39(3), 333–333 (2011).
- Fragoso-Bargas N , OpsahlJO , KiryushchenkoNet al. Cohort profile: Epigenetics in Pregnancy (EPIPREG) – population-based sample of European and South Asian pregnant women with epigenome-wide DNA methylation (850k) in peripheral blood leukocytes. PLoS One16(8), e0256158 (2021).
- Sobczyńska-Malefora A . Chapter 11 – methods for assessment of folate (vitamin B9). In: Laboratory Assessment of Vitamin Status.HarringtonD ( Ed.). Academic Press, London, UK, 219–264 (2019).
- Oslo universitetssykehus . Vitamin B9 (Folsyre, folat). In: Brukkerhåndbok i medisinsk biokjemi OUS.OUS, Oslo, Norway (2021).
- Kinnunen TI , SletnerL , SommerCet al. Ethnic differences in folic acid supplement use in a population-based cohort of pregnant women in Norway. BMC Pregnancy Childbirth17(1), 143 (2017).
- Morkrid K , JenumAK , SletnerLet al. Failure to increase insulin secretory capacity during pregnancy-induced insulin resistance is associated with ethnicity and gestational diabetes. Eur. J. Endocrinol.167(4), 579–588 (2012).
- Friedewald WT , LevyRI , FredricksonDS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin. Chem.18(6), 499–502 (1972).
- Waage CW , MdalaI , StigumHet al. Lipid and lipoprotein concentrations during pregnancy and associations with ethnicity. BMC Pregnancy Childbirth.22(1), 246 (2022).
- Miller SA , DykesDD , PoleskyHF. A simple salting out procedure for extracting DNA from human nucleated cells. NucleicAcids Res.16(3), 1215 ( 1988).
- Min JL , HemaniG , SmithGDet al. Meffil: efficient normalization and analysis of very large DNA methylation datasets. Bioinformatics34(23), 3983–3989 (2018).
- Houseman EA , AccomandoWP , KoestlerDCet al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinform.13, 86 (2012).
- Pidsley R , ZotenkoE , PetersTJet al. Critical evaluation of the Illumina Methylation EPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol.17(1), 208 (2016).
- Chang CC , ChowCC , TellierLCAMet al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience4, 7 (2015).
- Altshuler DM , DurbinRM , AbecasisGRet al. A global reference for human genetic variation. Nature526(7571), 68 (2015).
- Howie BN , DonnellyP , MarchiniJ. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet.5(6), e1000529 (2009).
- Du P , ZhangXA , HuangCCet al. Comparison of beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform.11, 587 (2010).
- Kuznetsova A , BrockhoffPB , ChristensenRHB. lmerTest package: tests in linear mixed effects models. J. Stat. Softw.82(13), 1–26 (2017).
- Ritchie ME , PhipsonB , WuDet al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res.43(7), e47 (2015).
- Liao YX , WangJ , JaehnigEJ , ShiZA , ZhangB. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res.47(W1), W199–W205 (2019).
- Pan H , HolbrookJD , KarnaniN , KwohCK. Gene, Environment and Methylation (GEM): a tool suite to efficiently navigate large scale epigenome wide association studies and integrate genotype and interaction between genotype and environment. BMC Bioinform.17, 299 (2016).
- Machiela MJ , ChanockSJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics31(21), 3555–3557 (2015).
- Tingley D , YamamotoT , HiroseKet al. mediation: R Package for causal mediation analysis. J. Stat. Softw.59(5), 1–38 (2014).
- Magnus P , BirkeC , VejrupKet al. Cohort profile update: the Norwegian Mother and Child Cohort Study (MoBa). Int. J. Epidemiol.45(2), 382–388 (2016).
- Paltiel L , AnitaH , SkjerdenTet al. The biobank of the Norwegian Mother and Child Cohort Study – present status. Nor. Epidemiol.24(1-2), 29–35 (2014).
- Battram T , YousefiP , CrawfordGet al. The EWAS Catalog: a database of epigenome-wide association studies [version 2; peer review: 2 approved]. Wellcome Open Res.7, 41 (2022).
- Gaunt TR , ShihabHA , HemaniGet al. Systematic identification of genetic influences on methylation across the human life course. Genome Biol.17, 61 (2016).
- Min JL , HemaniG , HannonEet al. Genomic and phenotypic insights from an atlas of genetic effects on DNA methylation. Nat. Genet.53(9), 1311–1321 (2021).
- Kamat MA , BlackshawJA , YoungRet al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics35(22), 4851–4853 (2019).
- Baric IC , SatalicZ , PedisicZet al. Validation of the folate food frequency questionnaire in vegetarians. Int. J. Food. Sci. Nutr.60, 88–95 (2009).
- Brantsaeter AL , HaugenM , AlexanderJ , MeltzerHM. Validity of a new food frequency questionnaire for pregnant women in the Norwegian Mother and Child Cohort Study (MoBa). Matern. Child. Nutr.4(1), 28–43 (2008).
- Zekovic M , Djekic-IvankovicM , NikolicMet al. Validity of the food frequency questionnaire assessing the folate intake in women of reproductive age living in a country without food fortification: application of the method of triads. Nutrients9(2), 128 (2017).
- Hackett A . Food frequency questionnaires: simple and cheap, but are they valid?Matern. Child Nutr.7(2), 109–111 (2011).
- Chen YY , GuptaMB , GratttonRet al. Down-regulation of placental folate transporters in intrauterine growth restriction. J. Nutr. Biochem.59, 136–141 (2018).
- Suliburska J , KocylowskiR , GrzesiakMet al. Evaluation of folate concentration in amniotic fluid and maternal and umbilical cord blood during labor. Arch. Med. Sci.15(6), 1425–1432 (2019).
- Yun HD , VarmaA , HussainMJet al. Clinical relevance of immunobiology in umbilical cord blood transplantation. J. Clin. Med.8(11), 1968 (2019).
- Sleven H , WelshSJ , YuJet al. De novo mutations in EBF3 cause a neurodevelopmental syndrome. Am. J. Hum. Genet.100(1), 138–150 (2017).
- Chao HT , DavidsM , BurkeEet al. A syndromic neurodevelopmental disorder caused by de novo variants in EBF3. Am. J. Hum. Genet.100(1), 128–137 (2017).
- Padhi EM , HayeckTJ , ChengZet al. Coding and noncoding variants in EBF3 are involved in HADDS and simplex autism. Hum. Genomics15(1), 44 (2021).
- Tao YF , XuLX , LuJet al. Early B-cell factor 3 (EBF3) is a novel tumor suppressor gene with promoter hypermethylation in pediatric acute myeloid leukemia. J. Exp. Clin. Cancer Res.34, 4 (2015).
- Rodger EJ , ChatterjeeA , StockwellPA , EcclesMR. Characterisation of DNA methylation changes in EBF3 and TBC1D16 associated with tumour progression and metastasis in multiple cancer types. Clin. Epigenetics11(1), 114 (2019).
- Coit P , RoopnarinesinghX , Ortiz-FernandezLet al. Hypomethylation of miR-17-92 cluster in lupus T cells and no significant role for genetic factors in the lupus-associated DNA methylation signature. Ann. Rheum. Dis.81(10), 1428–1437 (2022).
- Hochrainer K , PejanovicN , OlaseunVA , ZhangSet al. The ubiquitin ligase HERC3 attenuates NF-κB-dependent transcription independently of its enzymatic activity by delivering the RelA subunit for degradation. Nucleic Acids Res.43(20), 9889–9904 (2015).
- Mikkelsen K , ApostolopoulosV. Vitamin B12, folic acid, and the immune system. In: Nutrition and Immunity.MahmoudiM, RezaeiN ( Eds). Springer International Publishing, Cham, Switzerland, 103–114 (2019).
- McRae MP . High-dose folic acid supplementation effects on endothelial function and blood pressure in hypertensive patients: a meta-analysis of randomized controlled clinical trials. J. Chiropr. Med.8(1), 15–24 (2009).
- Shen MX , TanHZ , ZhouSJet al. Serum folate shows an inverse association with blood pressure in a cohort of chinese women of childbearing age: a cross-sectional study. PLoS One11(5), e0155801 (2016).
- Liu C , LiuCD , WangQS , ZhangZY. Supplementation of folic acid in pregnancy and the risk of preeclampsia and gestational hypertension: a meta-analysis. Arch. Gynecol. Obstet.298(4), 697–704 (2018).
- Wang Y , ZhaoN , QiuJet al. Folic acid supplementation and dietary folate intake, and risk of preeclampsia. Eur. J. Clin. Nutr.69(10), 1145–1150 (2015).
- Yadav S , LongkumerI , JoshiS , SaraswathyKN. Methylenetetrahydrofolate reductase gene polymorphism, global DNA methylation and blood pressure: a population based study from North India. BMC Med. Genomics14(1), 59 (2021).
- Pescador-Tapia A , Silva-MartinezGA , Fragoso-BargasNet al. Distinct associations of BMI and fatty acids with DNA methylation in fasting and postprandial states in men. Front. Genet.12, 665769 (2021).
- Grimes SB , WildR. Effect of pregnancy on lipid metabolism and lipoprotein levels. In: Endotext [Internet]. FeingoldKR, AnawaltB, BoyceAet al.et al. ( Eds). MDText.com, Inc., MA, USA (2000).