Abstract
Background: Necrotizing enterocolitis (NEC) is the most common gastrointestinal emergency in preterm infants. Epigenetic changes in DNA methylation may be present prior to NEC onset. Methods: 24 preterm infants with NEC and 45 matched controls were included. Human DNA was isolated from stool samples and methylation of CTDSPL2, HERC1, NXPE3 and PTGDR was measured using pyrosequencing. Results:CTDSPL2 displayed a higher DNA methylation of 51% compared with 17% in controls, prior to NEC onset (p = 0.047). Discussion: Noninvasive measurement of methylation in stool allows for comparison with healthy preterm controls. This potentially allows future biomarker or risk predictor use. The effect of CTDSPL2 hypermethylation on gene expression remains unclear.
Plain language summary
What is this article about?
Necrotizing enterocolitis (NEC) is a common emergency condition affecting the gastrointestinal system of preterm infants. Epigenetic changes in DNA methylation may be present in infants before the onset of NEC. DNA methylation is a natural process that can help turn genes on or off, thereby affecting their function. This study focused on measuring the amount of DNA methylation in certain genes in preterm infants who developed NEC.
What were the results?
This study included 24 preterm infants with NEC and 45 matched healthy controls. The researchers isolated human DNA from stool samples, and the amount of DNA methylation of four specific genes was measured. They found that one of the genes, CTDSPL2, had significantly higher DNA methylation in infants who later developed NEC than in healthy infants.
What do the results of the study mean?
In this study, researchers found that CTDSPL2 showed a higher level of DNA methylation in stool samples of infants who later developed NEC. The effect of this change remains unclear, but may affect the way cells grow and respond to injury or infection, which could contribute to the development of NEC. Measuring DNA methylation in stool samples provides a noninvasive method for identifying DNA methylation changes in preterm infants. Comparing the amount of DNA methylation in healthy infants with that in preterm infants at risk of NEC may help predict the risk of developing NEC.
Tweetable abstract
DNA samples from infants show higher DNA methylation of the marker gene CTDSPL2 prior to necrotizing enterocolitis #NEC onset.
Necrotizing enterocolitis (NEC) is the most common gastrointestinal emergency in preterm infants, characterized by inflammation of the intestine. It primarily affects preterm neonates, with the peak incidence occurring at around 31 weeks of postconceptional age [Citation1]. The condition can progress rapidly, with subtle symptoms escalating to a critical state that can ultimately lead to death [Citation2]. Despite advances in medical care, the mortality rate of infants with NEC has remained stagnant over the past three decades, with approximately 30% of cases resulting in death [Citation3]. The most important risk factor for developing NEC is prematurity, infants with the lowest gestational age at birth have the highest risk of developing NEC [Citation4]. Additional risk factors include alterations in intestinal microbiota colonization, hypoxic-ischemic injury, antibiotic use and formula feeding [Citation5–8]. However, not all infants with a similar risk profile develop NEC, leaving the question of why some develop NEC and others do not [Citation9]. Considering the unchanged survival rate, incomplete risk profile and devastating effect of this disease on patients and their families [Citation10], there is an urgent need for a better understanding of its pathophysiology, as well as an early detection method.
Alterations in DNA methylation of CpGs, especially in the promoter region of a gene, affect gene expression [Citation11]. The fetal and neonatal periods are critical to establish a stable DNA methylation pattern, and numerous CpGs with differential methylation have been identified in preterm born infants [Citation12]. These changes may influence the microbiome and immune processes, ultimately affecting susceptibility to NEC development [Citation13,Citation14]. In addition, changes in the microbiome may lead to changes in DNA methylation [Citation15]. DNA methylation may also serve to improve diagnostic accuracy of a disease, and a panel of DNA methylation changes has been used to improve early lung and colorectal cancer detection [Citation16,Citation17]. Recently, the current authors identified an altered methylome signature, including changes in TLR4 methylation, prior to NEC, corresponding to an altered immunological function predisposing infants to developing NEC [Citation18]. TLR4 plays a critical role in NEC development, as the starting point of a heightened inflammatory response after activation by lipopolysaccharide (LPS) on the bacterial cell wall [Citation19,Citation20]. A study of preterm-born pigs affected by NEC underlines the variable and dynamic nature of DNA methylation in the period after birth, which was strongly influenced by exposing the immature intestine to formula feeding [Citation21]. In infants with surgically treated NEC, other investigators have observed global hypermethylation in both colon and intestinal tissue after surgical resection, resulting in changes in gene expression [Citation22]. They observed a similar pattern in intestinal cells derived from stool samples of infants with NEC, after NEC onset [Citation23].
In the current study, stool samples obtained prior to NEC onset were used to determine the DNA methylation pattern of four prominent genes reported to be commonly hypermethylated in surgical NEC specimens. This offers a unique opportunity to detect whether these changes in methylation are already present prior to NEC onset and may play a role in NEC susceptibility rather than an effect of NEC itself. In addition, noninvasive measurement of DNA methylation using stool samples could be used as an early detection method for NEC.
Methods
Infants from a previously described case-control cohort were included [Citation18]. All included infants were born after a gestation of less than 30 weeks. 24 infants in this cohort developed NEC, Bell’s stage 2 or higher. 45 infants in the cohort were included as controls and matched to the cases based on gestational age, birth weight and sex. For this study, residual stool samples that were collected daily from birth onward were used. These samples were obtained from a completed multicenter observational cohort study on fecal volatile organic compounds. For all infants included in the study, written informed consent was obtained from all parents. The study was approved by the Institutional Review Board of the University Medical Center Groningen and use of these residual stool samples was permitted under the Dutch Medical Research with Human Subjects Law.
Human DNA was isolated from stool samples and collected as close as possible to the onset of NEC. The median collection time was one day prior to NEC onset. Most of the human DNA found in stool samples is derived from intestinal epithelial cells that renew every few days and shed into the stool. In infants, the main cell types shed into the stool are enterocytes, goblet cells, Paneth cells and enteroendocrine cells, while only a small proportion of lymphoid cells are present [Citation24]. Sample selection and DNA isolation were performed as previously reported using the QIAamp Fast DNA Stool Mini Kit (Qiagen, Hilden, Germany) [Citation18]. After DNA isolation, samples were treated with bisulfite using the EZ-96 DNA methylation Gold Kit (Zymo Research, CA, USA), following the protocol provided by the supplier. Genes were selected for analysis from a previously published dataset that included differentially methylated CpG sites in NEC colon. In that study, DNA methylation was calculated from colonic tissue obtained during surgical treatment of NEC [Citation22]. All differentially methylated single CpG sites identified in the colon by Good et al. (in their Supplemental Table 4A) were ranked based on q-values [Citation22]. Genes were excluded if they were not expressed in colon tissue, were nonprotein-coding or if SNPs were present at the CpG site. The location of the identified CpG position within the gene (e.g., inside or outside the promoter region) was not considered. This resulted in the selection of four genes with a highly significant differentially methylated CpG site in surgically resected colon tissue from NEC patients: PTGDR, HERC1, NXPE3 and CTDSPL2. Primers for these genes were designed to include the significant CpG position reported [Citation22] using Pyromark Assay Design software (Qiagen, Hilden, Germany). For HERC1 and PTGDR, two additional CpG positions were included that were in close proximity to the originally reported CpG positions. The CTDSPL2 CpG position of interested is located on chromosome 15, starting at nucleotide 44429117. For HERC1, the CpG position of interest was located on chromosome 15, starting at nucleotide 63678136. The CpG position of interest for NXPE3 was located on chromosome 3, starting at nucleotide 101804213. For PTGDR, the CpG position of interest started at nucleotide 52272903 on chromosome 14.
In total, methylation of six CpG positions was analyzed for these four genes (). DNA amplification was done using 12.5 μl of Qiagen HotStar Taq Master Mix, 1 μl of 10 μM forward and reverse primer, 5 μl of bisulfite converted DNA and 6.5 μl purified water. Cycling conditions on a T100 Thermal Cycler (Bio-Rad, CA, USA) were as follows: 95°C for 15 min, 50 cycles of 94°C for 30 s, 54°C (for PTGDR and NXPE3 this step was performed at 52°C) for 30 s, 72°C for 30 s, followed by a final step of 72°C for 7 min and storage at 4°C. Due to the generally low yield of human DNA from stool samples, the PCR protocol was optimized by increasing the number of cycles to 50 to enhance DNA amplification. Methylation was measured by pyrosequencing using the Pyromark Q24 (Qiagen, Hilden, Germany), following the provided manufacturer’s protocol. Data was analyzed using PyroMark Q24 Advanced Software (Qiagen). Samples that failed quality control or displayed low peak height were excluded from the data analysis. For HERC1, because of the low yield after PCR, only samples with a visible band on gel electrophoresis were analyzed using pyrosequencing. Samples that showed low peak height during pyrosequencing due to low DNA content were analyzed separately but were not included in the current data analysis.
Table 1. DNA methylation of CTDSPL2, HERC1, NXPE3 and PTGDR.
Statistical analyses were conducted using IBM SPSS Statistics for Windows, version 28.0 (IBM Corp., NY, USA). A parametric or nonparametric approach was used based on the examination of Q-Q plots. Continuous variables were analyzed using either the Mann-Whitney U test or Student’s t-test. Correlations were assessed through visual inspection of scatterplots and further evaluated using Pearson’s test or Spearman’s Rho. p-values less than 0.05 (two-tailed) were considered statistically significant. Figures were constructed using R, version 4.2.3. (R Foundation for Statistical Computing, Vienna, Austria).
Results
A large variation in DNA methylation across all samples was observed at all CpG positions (). Only CTDSPL2 showed a significant difference in DNA methylation prior to NEC onset, with a 51% median methylation in infants with NEC compared with 17% in controls (p = 0.047; ). The variation in methylation of CTDSPL2 is also visualized in F. PTGDR CpG position 1 (A) showed a median methylation of 76.19% in infants with NEC compared with 78.49% in controls (p = 0.495), and position 2 (B) showed a median methylation of 77.35% versus 66.94% (p = 0.242). NXPE3 (C) displayed a median methylation of 81.26% in infants with NEC compared with 58.38% in controls (p = 0.582). For infants with NEC, the median methylation of HERC1 CpG position 1 (D) was 100% versus 98.53% in controls (p = 0.815), and for CpG position 2 (E) this was 95.72% compared with 81.26% in controls (p = 0.743). The DNA methylation of all tested genes did not correlate significantly with gestational age, birth weight, NEC severity or postnatal age at stool sample collection (). Due to the lower quality of human DNA isolated from stool samples, many of the samples did not pass the strict quality control checks after pyrosequencing, resulting in a lower number of included infants. The main clinical characteristics for infants included in the analysis for CTDSPL2 are presented in Supplementary Table 1. After excluding missing samples for this gene, no significant differences were found in gestational age, birthweight or sex between the NEC and control groups.
Boxes (A–F) show the DNA methylation of PTGDR, NXPE, HERC1 and CTDSPL respectively. *Only CTDSPL2 showed a significant difference in DNA methylation prior to NEC onset, with a 51% median methylation in infants with NEC compared with 17% in controls; p = 0.047.
NEC: Necrotizing enterocolitis.
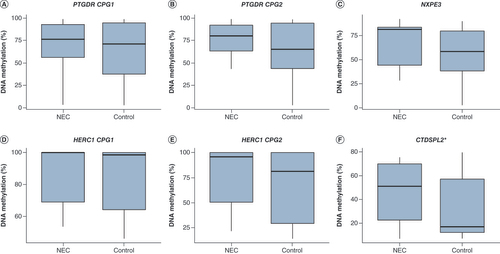
Table 2. Absence of association between DNA methylation of CTDSPL2, HERC1, NXPE3 and PTGDR and clinical characteristics.
To investigate whether the methylation of these genes was affected by NEC or a precursor of NEC, whether the collection date of the stool samples in relation to NEC onset was related to the percentage of DNA methylation was assessed. No correlation was found between the exact number of days before NEC onset and DNA methylation ().
Discussion
DNA methylation may play a role in predisposing infants to NEC. Of four previously identified hypermethylated genes in patients who already developed NEC [Citation22], only CTDSPL2 displayed significantly higher DNA methylation prior to NEC onset. At the same CpG position as previously identified by Good et al. (1487 nucleotides after the transcription start site), our findings demonstrated a methylation difference of 34 percentage points when comparing samples taken prior to NEC onset with those of healthy preterm age-matched controls. Good et al. demonstrated hypermethylation of this gene after NEC onset, with a methylation difference of 47 percentage points when comparing infants with surgical NEC to controls [Citation4]. Their control group consisted of an unmatched group of infants who had either healed from NEC and underwent an operation for reanastomosis or infants with a spontaneous intestinal perforation or anorectal malformation. In a follow-up study using stool samples, general hypermethylation was reported, although CTDSPL2 was not mentioned. In the latter study, a small number of infants were included and stool samples were collected after NEC onset [Citation23].
CTDSPL2, also known as SCP4, has a role in regulating cell mitosis, proliferation and motility. CTDSPL2 plays a role in pancreatic tumors and B-cell lymphoma because its overexpression promotes cell migration and immortalization [Citation25,Citation26]. Hypermethylation of CTDSPL2 may result in reduced expression, leading to CTDSPL2 depletion. This may restrict cell cycle progression and induce inflammation through the NF-ĸB pathway [Citation26]. This pathway is also utilized by TLR4 to induce an exaggerated inflammatory response, as observed in NEC [Citation20,Citation27]. Thus, CTDSPL2 depletion may contribute to NEC onset. Only one other study has reported the methylation of CTDSPL2, in a cohort of obese asthmatic preadolescent children [Citation28]. In this study, hypermethylation of CTDSPL2 was reported in cases compared with controls, but gene expression was not measured.
The current results demonstrated that hypermethylation of a previously identified CpG site in CTDSPL2 is already present in human DNA derived from stool samples prior to the onset of NEC. We did not find any differences in the other three genes defined based on published data [Citation22]. Changes in DNA methylation in these genes may not yet be present prior to NEC or may be present only in severe surgical NEC cases. In our dataset, we could not confirm the latter because we did not find a correlation between DNA methylation and NEC severity. Alternatively, changes in DNA methylation may be more subtle prior to NEC and therefore not significantly different in this small cohort. Lastly, the studies by Good et al. [Citation22,Citation23] were specifically performed on epithelial cells, whereas our samples contain human DNA from various cell types. A limitation of the current study was the low quality of human DNA isolated from stool. Despite including a second or third sample to optimize DNA isolation from the included infants to reduce missing data, we still lost over 50% of the included infants for some genes after pyrosequencing. Other investigators report increasing amplification cycles as was done in this study as well, to increase DNA yield from stool samples for analysis [Citation23]. These missing infants in the study are a direct result of the decision to adhere to robust quality control criteria, which excluded samples that displayed low peak height during pyrosequencing, to ensure the reliability and accuracy of our data. This decision led to a reduction in the number of valid samples analyzed and impacted the statistical power of this study. The samples with low peak heights, identified after pyrosequencing, were analyzed separately and showed similar results to the included samples of high quality, but were not included in this analysis (data available upon request).
Finally, we note that there is a large overlap in methylation levels at the CTDSPL2 CpG site (F), which makes it difficult to distinguish between NEC cases and controls based on DNA methylation alone. The CTDSPL2 CpG site in this study is just one of many CpG sites that could potentially be used as a biomarker for NEC. Further research is needed to identify a potential panel of CpG sites that are specific to NEC and have less overlap in methylation levels between cases and controls. Especially, the promotor region of the gene would be of interest for these future investigations, as this region is heavily involved in regulating gene expression.
The effect of methylation changes in CTDSPL2 on the differences in gene expression remains unclear, since the studied CpG position of CTDSPL2 is not present in its promoter region. No previous studies have evaluated the effect of changes in DNA methylation of CTDSPL2 on gene expression in the intestine. Analysis of mRNA might be helpful, but these stool samples were not suitable for such analyses. Stool was collected at routine diaper changes to not disturb the infants and may have been in the diaper for several hours prior to transfer to the freezer. As mRNA is not likely to remain stable for this amount of time at room temperature, we were concerned about the reliability of any data obtained from such samples [Citation29]. In future studies, stool frozen immediately would be suitable for mRNA analysis using recent techniques such as droplet digital PCR [Citation30].
Conclusion
Significant hypermethylation of CTDSPL2 was found prior to NEC onset. Stool samples enabled a comparison between preterm-born infants with NEC and a matched control group without subsequent NEC, which is impossible when surgically resected material is used. In addition, the use of stool samples is the only noninvasive method currently available to measure DNA methylation in human DNA predominantly from epithelial cells of the intestine, of preterm-born infants prior to NEC onset. Future research is needed for a broader evaluation of CTDSPL2 DNA methylation to evaluate its role in NEC development beyond a possible role as biomarker or risk predictor. This research should include longitudinally collected stool samples prior to NEC, combined with surgically obtained samples or freshly frozen stool samples, which would allow for expression studies.
In infants with surgically treated necrotizing enterocolitis (NEC), investigators have observed global hypermethylation in both colon and intestinal tissue after surgical resection.
This is the first study evaluating those changes in DNA methylation prior to the onset of NEC and including healthy controls.
CTDSPL2 displayed a higher DNA methylation of 51% compared with 17% in controls, prior to NEC onset (p = 0.047).
PTGDR, HERC1 and NXPE3 did not show significant differences in DNA methylation when comparing cases to controls, prior to NEC onset.
DNA methylation of all tested genes did not correlate significantly with gestational age, birth weight, NEC severity, time to NEC onset or postnatal age at stool sample collection.
The effects of these methylation changes on the differences in gene expression remain unclear, since the studied CpG position of CTDSPL2 is not present in the promoter region.
Noninvasive measurement of methylation using stool samples allows for comparison with healthy preterm controls and potential biomarker or risk predictor use.
Future research is needed for a broader evaluation of CTDSPL2 DNA methylation to evaluate its role in NEC development, or as a possible biomarker or risk predictor.
D Klerk, T Plösch, J Hulscher, E Kooi and A Bos conceptualized and designed the study. D Klerk and R Verkaik-Schakel collected and analyzed the data. D Klerk drafted the initial manuscript. T Plösch, E Kooi and A Bos supervised the study. R Verkaik-Schakel, T Plösch, J Hulscher, E Kooi and A Bos critically reviewed and revised the manuscript. All authors approved the final manuscript as submitted.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Supplemental Table
Download MS Word (13.6 KB)Acknowledgments
The authors would like to thank the nurses and medical staff of the NICU of the Beatrix Children’s Hospital in Groningen, as well as the laboratory staff of the obstetrics laboratory for helping with data collection and for creating the opportunity to carry out this study.
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.tandfonline.com/doi/suppl/10.2217/epi-2023-0119
Financial & competing interests disclosure
A research grant was obtained for this work from the Dutch de Cock-Hadders Foundation. D Klerk was financially supported by the Junior Scientific Masterclass of the University of Groningen. The open access article processing charges were paid through the University of Groningen open access fund. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Data sharing statement
The datasets analyzed in the current study, including deidentified participant data, are available immediately after publication, upon reasonable request. Proposals may be emailed to the corresponding author and include a methodological and sound proposal. Data requesters must sign a data access agreement.
Additional information
Funding
References
- Neu J . Necrotizing enterocolitis. Semin. Fetal Neonatal Med.23(6), 369 (2018).
- Zani A , PierroA. Necrotizing enterocolitis: controversies and challenges. F1000Res4, 1373 (2015).
- Heida FH , StolwijkL , LoosMHet al. Increased incidence of necrotizing enterocolitis in the Netherlands after implementation of the new Dutch guideline for active treatment in extremely preterm infants: results from three academic referral centers. J. Pediatr. Surg.52(2), 273–276 (2017).
- Lin PW , StollBJ. Necrotising enterocolitis. Lancet368(9543), 1271–1283 (2006).
- Singh DK , MillerCM , OrgelKA , DaveM , MackayS , GoodM. Necrotizing enterocolitis: bench to bedside approaches and advancing our understanding of disease pathogenesis. Front. Pediatr.10, 1107404 (2022).
- Rina P , ZengY , YingJ , QuY , MuD. Association of initial empirical antibiotic therapy with increased risk of necrotizing enterocolitis. Eur. J. Pediatr.179(7), 1047–1056 (2020).
- Warner BB , DeychE , ZhouYet al. Gut bacteria dysbiosis and necrotising enterocolitis in very low birthweight infants: a prospective case-control study. Lancet387(10031), 1928–1936 (2016).
- Kaplina A , KononovaS , ZaikovaE , PervuninaT , PetrovaN , SitkinS. Necrotizing enterocolitis: the role of hypoxia, gut microbiome, and microbial metabolites. Int. J. Mol. Sci.24(3), 2471 (2023).
- Samuels N , VanDe Graaf RA , DeJonge RCJ , ReissIKM , VermeulenMJ. Risk factors for necrotizing enterocolitis in neonates: a systematic review of prognostic studies. BMC Pediatr.17(1), 105 (2017).
- Canvasser J , PatelRM , PryorEet al. Long-term outcomes and life-impacts of necrotizing enterocolitis: a survey of survivors and parents. Semin. Perinatol.47(1), 151696 (2023).
- Greenberg MVC , Bourc’hisD. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol.20(10), 590–607 (2019).
- Merid SK , NovoloacaA , SharpGCet al. Epigenome-wide meta-analysis of blood DNA methylation in newborns and children identifies numerous loci related to gestational age. Genome Med.12(1), 25 (2020).
- Cortese R , LuL , YuY , RudenD , ClaudEC. Epigenome-microbiome crosstalk: a potential new paradigm influencing neonatal susceptibility to disease. Epigenetics11(3), 205–215 (2016).
- Lu L , FanJ , XuWet al. DNA methylome mapping identifies epigenetic abnormalities in intestinal lymphocyte regulation in human necrotizing enterocolitis. Dig. Dis. Sci.67(9), 4434–4443 (2022).
- Ansari I , RaddatzG , GutekunstJet al. The microbiota programs DNA methylation to control intestinal homeostasis and inflammation. Nat. Microbiol.5(4), 610–619 (2020).
- Raut JR , GuanZ , Schrotz-KingP , BrennerH. Fecal DNA methylation markers for detecting stages of colorectal cancer and its precursors: a systematic review. Clin. Epigenetics12(1), 122 (2020).
- Wei B , WuF , XingWet al. A panel of DNA methylation biomarkers for detection and improving diagnostic efficiency of lung cancer. Sci. Rep.11(1), 16782 (2021).
- Klerk DH , PloschT , Verkaik-SchakelRN , HulscherJBF , KooiEMW , BosAF. DNA methylation of TLR4, VEGFA, and DEFA5 is associated with necrotizing enterocolitis in preterm infants. Front. Pediatr.9, 630817 (2021).
- Hackam DJ , SodhiCP. Bench to bedside–new insights into the pathogenesis of necrotizing enterocolitis. Nat. Rev. Gastroenterol. Hepatol.19(7), 468–479 (2022).
- Hackam DJ , SodhiCP. Toll-like receptor-mediated intestinal inflammatory imbalance in the pathogenesis of necrotizing enterocolitis. Cell Mol. Gastroenterol. Hepatol.6(2), 229–238 (2018).
- Gao F , ZhangJ , JiangPet al. Marked methylation changes in intestinal genes during the perinatal period of preterm neonates. BMC Genomics15(1), 716 (2014).
- Good M , ChuT , ShawPet al. Global hypermethylation of intestinal epithelial cells is a hallmark feature of neonatal surgical necrotizing enterocolitis. Clin. Epigenetics12(1), 190 (2020).
- Good M , ChuT , ShawPet al. Neonatal necrotizing enterocolitis-associated DNA methylation signatures in the colon are evident in stool samples of affected individuals. Epigenomics13(11), 829–844 (2021).
- Chapkin RS , ZhaoC , IvanovIet al. Noninvasive stool-based detection of infant gastrointestinal development using gene expression profiles from exfoliated epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol.298(5), G582–589 (2010).
- Winans S , FlynnA , MalhotraS , BalagopalV , BeemonKL. Integration of ALV into CTDSPL and CTDSPL2 genes in B-cell lymphomas promotes cell immortalization, migration and survival. Oncotarget8(34), 57302–57315 (2017).
- Xiao Y , ChenY , PengA , DongJ. The phosphatase CTDSPL2 is phosphorylated in mitosis and a target for restraining tumor growth and motility in pancreatic cancer. Cancer Lett.526, 53–65 (2022).
- Mi Y , XieX , BaoZ , XiongX , WangX , ZhangH. Dimethyl fumarate protects against intestine damage in necrotizing enterocolitis by inhibiting the toll-like receptor (TLR) inflammatory signaling pathway. Tissue Cell81, 102003 (2023).
- Rastogi D , SuzukiM , GreallyJM. Differential epigenome-wide DNA methylation patterns in childhood obesity-associated asthma. Sci. Rep.3, 2164 (2013).
- Reck M , TomaschJ , DengZet al. Stool metatranscriptomics: a technical guideline for mRNA stabilisation and isolation. BMC Genomics16(1), 494 (2015).
- Stauber J , ShaikhN , OrdizMI , TarrPI , ManaryMJ. Droplet digital PCR quantifies host inflammatory transcripts in feces reliably and reproducibly. Cell. Immunol.303, 43–49 (2016).
