Abstract
Aim: To investigate SIX4 in breast cancer. Methods: Publicly available online tools were used to analyze the expression, methylation and prognostic significance of SIX4 in breast cancer, as well as its immunohistochemistry. Results: High SIX4 levels were associated with low SIX4 promoter methylation, especially in estrogen receptor-positive breast cancer. Increased SIX4 was related to advanced stage and decreased immune infiltration. Gene set enrichment analysis found that the SIX4-correlated genes were enriched in transcriptional processing and immune response. Patients with high SIX4 expression tended to have poor survival, especially those with estrogen receptor-positive breast cancer. Conclusion: High SIX4 expression in breast cancer plays an oncogenic role, promoting the development of malignancies through suppressing the immune response, especially in luminal subtypes, and is associated with a low promoter methylation level.
Tweetable abstract
High SIX4 levels in breast cancer, especially estrogen receptor-positive patients, are associated with low SIX4 promoter methylation. High SIX4 is related to advanced stage, decreased immune infiltration and poor survival.
Graphical abstract
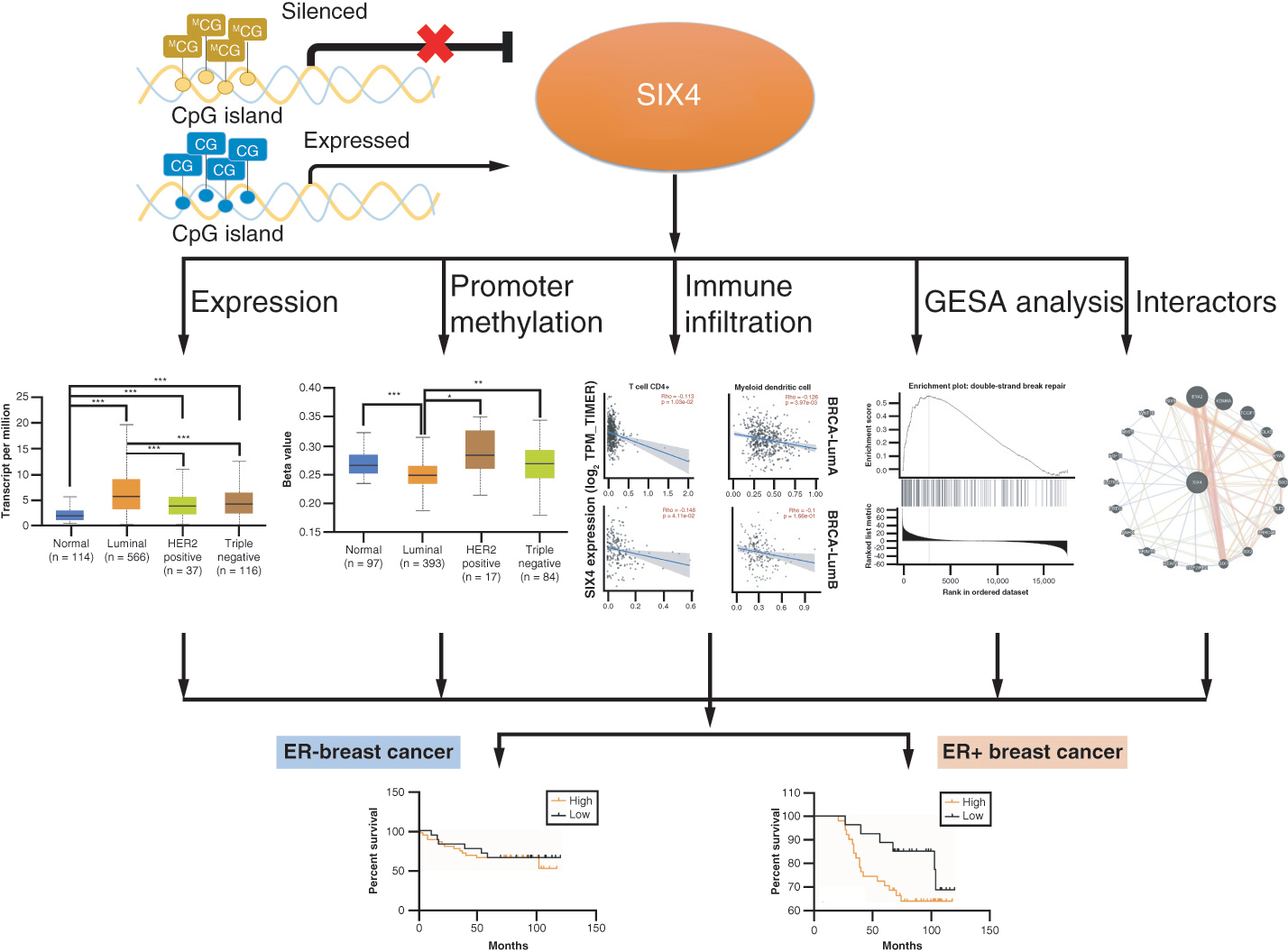
A schematic model depicting SIX4 upregulation in breast cancer, likely to be controlled by low SIX4 gene promoter methylation, especially in estrogen receptor-positive breast cancer.
The hallmarks of cancer, introduced by Hanahan and Weinberg, typify the essential phenotypes acquired by cancers and include limitless cell division, resistance to programmed cell death, initiation of invasion and metastasis and the induction of angiogenesis [Citation1]. However, the non-ideal prognosis of patients with malignancies indicates that the underlying molecular mechanisms are still not well understood. Breast cancer, the most diagnosed type of cancer affecting women, is also one of the most common causes of cancer-related death worldwide [Citation2] and can be divided into different histological and molecular subtypes with distinct clinical manifestations, treatment strategies, responses to therapy and clinical outcomes [Citation3]. Interestingly, immunotherapy has been of limited use despite breast cancer being an immunogenic tumor [Citation3]. Understanding the complex interactions between tumor and immune cells, and associated molecular mechanisms in subtypes of breast cancer, will provide a perspective for the applicability of immunotherapy for different subtypes.
Human SIX4, a member of the homeobox family, functions as a transcription factor to regulate the differentiation or maturation of neuronal cells [Citation4]. As a key regulator of organogenesis, especially in brain and craniofacial structures, SIX4 is unsurprisingly involved in the differentiation of embryonic stem cells to mediate organ development [Citation5–7]. Abnormal expression of SIX4 has been reported in gastric cancer [Citation8], ovarian cancer [Citation9], cervical cancer [Citation10], hepatocellular carcinoma [Citation11], non-small-cell lung cancer [Citation12], breast cancer [Citation13] and esophageal squamous cell carcinoma [Citation14]. Our previous study identified the special role of SIX4 in colorectal cancer (CRC), among different SIX family members, as a potential therapeutic target involved in oxidative phosphorylation and respiratory chain signaling pathways [Citation15]. Because it is an immunogenic disease, patients with breast cancer show diverse responses to specific therapeutic strategies based on the molecular type. However, the effects and potentially distinct mechanisms of the oncogenic role of SIX4 in different subtypes of breast cancer are unclear. The current study focuses on the prognostic significance of SIX4 in breast cancer, and epigenetic alteration associated with dysregulation of SIX4 expression in different breast cancer subtypes, in order to provide novel subtype-specific therapeutic targets for breast cancer.
Materials & methods
Expression of SIX4 in different types of malignancies
The expression of SIX4 in tumor and normal samples was analyzed using Gene Expression Profiling Interactive Analysis (GEPIA) 2 [Citation16]. GEPIA2 (http://gepia2.cancer-pku.cn/#index) is a valuable and highly cited resource for gene expression analysis of tumor and normal samples, and has a total of 33 types of human tumors, from The Cancer Genome Atlas (TCGA) and the Genotype–Tissue Expression (GTEx) database. To assess the protein level of SIX4 in different types of malignancies, the Human Protein Atlas (www.proteinatlas.org/) was applied [Citation17].
Correlation analysis between SIX4 & clinicopathological parameters in breast cancers
To investigate the potential function of high SIX4 levels in breast cancer, the University of Alabama at Birmingham Cancer (UALCAN) portal (https://ualcan.path.uab.edu/) was used to investigate the relationship between SIX4 level and clinicopathological parameters in patients with breast cancer [Citation18]. UALCAN is an online tool for analyzing specific gene levels from TCGA patient cohorts, including 1097 primary breast cancers and 114 normal samples. The stage of tumor, lymph node metastasis and molecular classification were used for the clinicopathological parameters for further analysis using UALCAN. The methylation level of promoter regions was also evaluated through UALCAN.
Prognostic analysis of SIX4 expression in patients with breast cancer
The prognostic value of SIX4 in patients with breast cancer was assessed in UALCAN, including overall survival (OS) and disease-free survival (DFS). A total of 2032 breast cancer samples in the Kaplan–Meier Plotter (http://kmplot.com/analysis/) database [Citation19] were used to predict the role of SIX4 levels in relapse-free survival (RFS) of breast cancer. Prediction Analysis of Microarray 50 (PAM50) was used for molecular subtyping of breast cancer patients into luminal A, luminal B, HER2+, basal-like and normal-like [Citation20]. Patients with breast cancer were divided into high and low SIX4 expression groups according to the median expression level of SIX4.
Mutation of SIX4 & SIX4-correlated genes in breast cancer
Breast invasive carcinoma (TCGA, PanCancer Atlas), with 1084 samples in the cBioPortal (http://kmplot.com/analysis/) platform [Citation21], was used to investigate the genetic alteration of SIX4 in breast cancer. To find the potential involvement of SIX4 in breast cancer, LinkedOmics and gene set enrichment analysis (GSEA) (www.linkedomics.org/) were applied to analyze the SIX4-correlated genes [Citation22]. The Pearson correlation test was used to evaluate 20,155 gene IDs in 1093 patients with breast cancer and draw their association with SIX4. Then GSEA was conducted to predict the involved SIX4-related signaling pathways. Normalized enrichment scores were calculated as the main parameter for enriching SIX4 in different gene sets. The cutoff was set as |normalized enrichment score|>1, false discovery rate <0.25 and p < 0.05. To verify the correlation of SIX4 with specific genes, the co-expression module in cBioPortal was applied based on Spearman analysis. A protein–protein interaction (PPI) network related to SIX4 was also constructed on GeneMANIA (https://genemania.org/), a flexible web interface [Citation23].
Immune infiltration related to SIX4 expression in breast cancers
Based on the GSEA findings, we focused on the immune response as related to the expression of SIX4 in breast cancer. The infiltration level of different immune cells was evaluated using TIMER 2.0 (http://timer.cistrome.org/), including B cells, CD4+/CD8+ T cells, myeloid dendritic cells, macrophages and neutrophils [Citation24]. TIMER 2.0 is an enhanced version to integrate multiple state-of-the-art algorithms (including TIMER, xCell, MCP-counter, CIBERSORT, EPIC and quanTIseq) for infiltration estimation, enabling users to visualize all the estimations together to reach more confident conclusions [Citation24].
Patient information & ethics statement
To evaluate the protein levels of SIX4 in patients with breast cancer, a tissue microarray with breast cancer tissues was obtained from OUTDO (Shanghai, China). All patients were females, with ages ranging from 37 to 88 years old. All patients were diagnosed at the first visit as having primary breast cancer, without evidence of metastasis. This study was approved by the Ethics Committee of Shantou University Medical College.
Immunohistochemical staining & evaluation
Deparaffinization in xylene, hydration in graded alcohols, epitope retrieval in ethylenediaminetetraacetic acid and blocking endogenous peroxidase in 3% H2O2 were performed on the tissue microarray as described previously [Citation25]. The slide was incubated with anti-SIX4 antibody at 1:400 dilution (bs-17503R, Bioss, Beijing, China) overnight, followed by hematoxylin nuclear counterstaining for visualization.
Two individual investigators without prior knowledge of patient information evaluated the immunohistochemistry staining results, based on the staining intensities and the percentage of positive cells. The final staining score of SIX4 expression in breast cancer tissues was calculated as the sum of staining intensity and percentage of positive cells, and divided into low- and high-expression groups.
Statistical analyses
The relationship between SIX4 level and clinicopathological parameters of breast cancer patients was analyzed using the χ-square or Fisher’s exact probability test. To evaluate the prognostic value of the protein level of SIX4 in breast cancer, Kaplan–Meier survival curves were used with the Gehan–Breslow–Wilcoxon test for p-value determination. Differences with p < 0.05 were considered to be statistically significant.
Results
SIX4 expression is high in breast cancer tissues compared with normal tissues
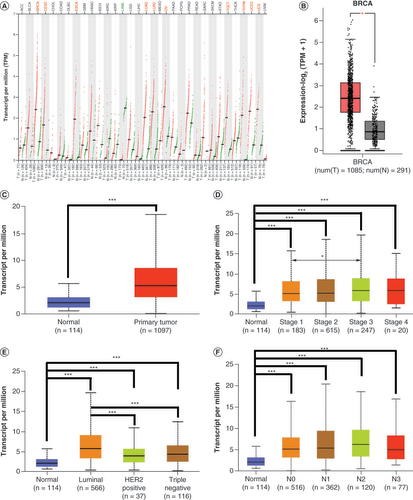
In different types of cancers, the expression levels of SIX4 were diverse (A). Compared with corresponding normal tissue, an increased level of SIX4 was found in breast invasive carcinoma, cervical squamous cell carcinoma and endocervical adenocarcinoma, esophageal carcinoma, lung adenocarcinoma, lung squamous cell carcinoma, ovarian serous cystadenocarcinoma, testicular germ cell tumor, thymoma, uterine corpus endometrial carcinoma and uterine carcinosarcoma tissues, whereas in non-solid malignancy and acute myeloid leukemia, the expression of SIX4 was decreased. The specific expression pattern of SIX4 in breast cancer compared to normal tissues is presented in B.
(A) Expression pattern of SIX4 in different types of malignancies. Dots in the figure represent different samples: red for tumors and green for normal tissue. In different types of malignancies, the cancerous tissues with significantly increased SIX4 are labeled in red and include BRCA, CESC, ESCA, LUAD, LUSC, OV, TGCT, THYM, UCEC and UCS, while malignant tissues with decreased SIX4 are labeled in green and include LAML. (B & C) The increased SIX4 level in breast cancer compared with normal tissues in GEPIA and UALCAN. (D) Expression of SIX4 in different stages of breast cancer. (E) Expression of SIX4 in different molecular subtypes of breast cancer. (F) Expression of SIX4 in different lymph node statuses.
BRCA: Breast invasive carcinoma; CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma; ESCA: Esophageal carcinoma; GEPIA: Gene Expression Profiling Interactive Analysis; LAML: Acute myeloid leukemia; LUAD: Lung adenocarcinoma; LUSC: Lung squamous cell carcinoma; OV: Ovarian serous cystadenocarcinoma; TGCT: Testicular germ cell tumor; THYM: Thymoma; UALCAN: University of Alabama at Birmingham Cancer portal; UCEC: Uterine corpus endometrial carcinoma; UCS: Uterine carcinosarcoma.
High SIX4 expression is associated with increased disease stage of breast cancers
The increased expression of SIX4 in breast cancer was confirmed by analyzing the breast cancer cohort in the UALCAN portal (C). In advanced stages, the expression level of SIX4 tended to be high (D). Interestingly, among the different molecular types of breast cancer, the level of SIX4 was highest in the luminal subtypes, although in the HER2+ and triple-negative types, SIX4 was also increased (E). Regarding lymph node status, SIX4 expression had no relationship with lymph node invasion (F).
High levels of SIX4 predict poor survival of patients with breast cancer, especially for luminal subtypes
To determine the potential role of increased SIX4 in breast cancer, the prognostic value was evaluated for OS, DFS and RFS. Breast cancer patients with high SIX4 levels tended to have poorer OS (A) and DFS (B), although the difference was not statistically significant (p> 0.05).
(A) Overall survival. (B) Disease-free survival. (C) Relapse-free survival (RFS) in all patients. (D) Luminal A subtype RFS. (E) Luminal B subtype RFS. (F) HER2+ subtype RFS. (G) Basal-like subtype RFS. (H) Normal-like subtype RFS. (I) ER− patient RFS. (J) ER+ patient RFS. (K) PR− patient RFS. (L) PR+ patient RFS. (M) HER2− patient RFS. (N) HER2+ patient RFS. (O) RFS for patients without lymph node metastasis. (P) RFS for patients with lymph node metastasis.
ER: Estrogen receptor; PR: Progesterone receptor.
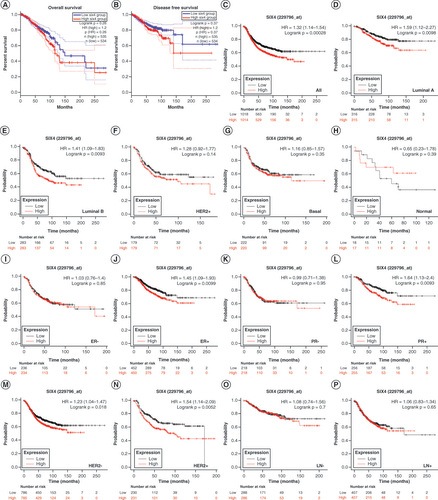
For further exploration, breast cancer relapse was analyzed. In all molecular types of breast cancer, a high level of SIX4 predicted a high rate of relapse in patients (C). Interestingly, in patients with luminal breast cancer, for both luminal A and luminal B, high expression of SIX4 was positively associated with poor RFS (D & E). However, in the other molecular types of breast cancer, no association was found between SIX4 level and RFS (F–H).
Based on the prognostic results of SIX4 for different molecular subtypes of breast cancer, the status of hormone receptors was determined for further analysis. Consistent with previous findings, in breast cancer patients who were both estrogen receptor-positive (ER+) and progesterone receptor-positive (PR+), SIX4 expression was positively related to poor survival and relapse (J & ), while no effect of SIX4 level on RFS was found in ER− or PR− patients (I & K). Regarding HER2 status, high SIX4 levels predicted poor RFS in both HER2− and HER2+ patients (M & N). However, no matter whether lymph node metastasis had or had not occurred, SIX4 expression did not affect RFS (O & P).
SIX4 gene promoter methylation is negatively related to its expression in breast cancer
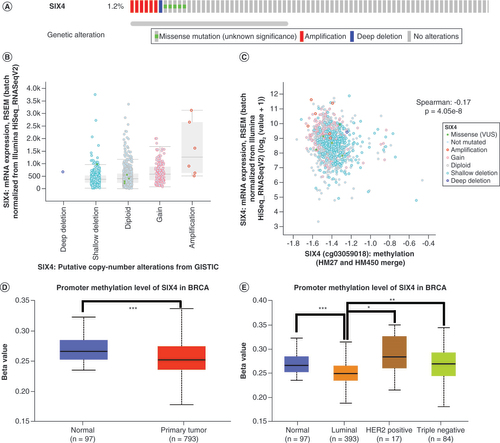
To explore the mechanism of increased SIX4 expression in breast cancer, the genetic alteration of SIX4 was analyzed using cBioPortal, revealing that most patients had no mutations in the SIX4 gene (A) and only a few had amplifications (B). Interestingly, the expression level of SIX4 was negatively related to its methylation, with r = -0.17 and p = 4.05e–8 in Spearman analysis (C). After comparing SIX4 gene promoter methylation in 97 normal and 793 primary breast cancer tissues using UALCAN, a decreased promoter methylation level was found in primary breast cancer, which was inversely related to SIX4 expression in breast cancer (D). Further in different molecular subtypes, the level of SIX4 gene promoter methylation was lowest in the luminal types of breast cancer (E), which had the highest expression of SIX4 (D), suggesting that the low promoter methylation likely contributed to increase the expression of SIX4 in breast cancer.
(A) Mutation percentage and types of SIX4 in breast cancer patients from The Cancer Genome Atlas, PanCancer Atlas. (B) Distribution of SIX4 expression in different copy number variation groups. (C) Correlation of SIX4 mRNA expression with copy number variation groups. (D) Promoter methylation level of SIX4 in primary breast cancer compared with normal breast tissue samples. (E) Promoter methylation level of SIX4 in different molecular subtypes of breast cancer.
SIX4 is enriched in transcriptional processes & immune response in breast cancer
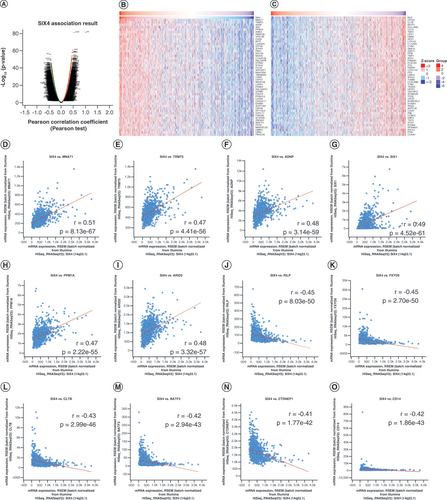
To elucidate the potential molecular mechanism of SIX4 involvement in breast cancer, LinkedOmics was used to search the breast invasive carcinoma dataset in TCGA for SIX4-related genes [Citation22]. A shows the association of different genes with SIX4 and their Pearson correlation coefficients. The top 50 positively correlated and top 50 negatively correlated genes are listed as a heat map in B & . To verify the top genes associated with SIX4, cBioPortal was applied and revealed that the positively associated genes were MNAT1 (r = 0.51; p = 8.13e-67; D), TRMT5 (r = 0.47; p = 4.41e-56; E), ADNP (r = 0.48; p = 3.14e-59; F), SIX1 (r = 0.49; p = 4.52e-61; G), PPM1A (r = 0.47; p = 2.22e-55; H) and ARID2 (r = 0.48; p = 3.32e-57; I), while negatively associated genes were RILP (r = -0.45; p = 8.03e-50; J), FXYD5 (r = -0.45; p = 2.70e-50; K), CLTB (r = -0.43; p = 2.99e-46; L), BATF3 (r = -0.42; p = 2.94e-43; M), DULLARD (CTDNEP1, r = -0.41; p = 1.77e-42; N) and CD14 (r = -0.42; p = 1.86e-43; O).
(A) Genes whose expression is correlated with that of SIX4 in breast cancer. (B) Top 50 genes positively correlated with SIX4. (C) Top 50 genes negatively correlated with SIX4. (D)MNAT1. (E)TRMT5. (F)ADNP. (G)SIX1. (H)PPM1A. (I)ARID2. (J)RILP. (K)FXYD5. (L)CLTB. (M)BATF3. (N)CTDNEP1 (DULLARD). (O)CD14.
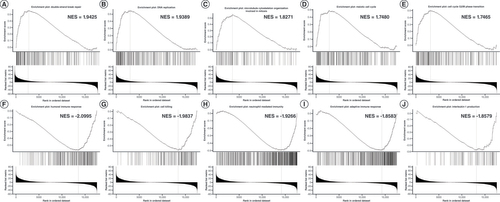
Next, GSEA was conducted for analyzing the enriched gene sets associated with SIX4. Interestingly, the associated genes were enriched in biological processes related to transcription and promotion of the cell cycle, such as double-strand break repair, DNA replication, microtubule cytoskeleton organization involved in mitosis, meiotic cell cycle and G2/M phase transition (A–E), indicating that SIX4 may enhance cell cycle processes as well as transcriptional activity. At the same time, enrichment of SIX4 was found in the processes of humoral immune response, cell killing, neutrophil-mediated immunity, adaptive immune response and IL-1 production (F–J).
(A) Double-strand break repair. (B) DNA replication. (C) Microtubule cytoskeleton organization involved in mitosis. (D) Meiotic cell cycle. (E) Cell cycle G2/M phase transition. (F) Humoral immune response. (G) Cell killing. (H) Neutrophil-mediated immunity. (I) Adaptive immune response. (J) IL-1 production.
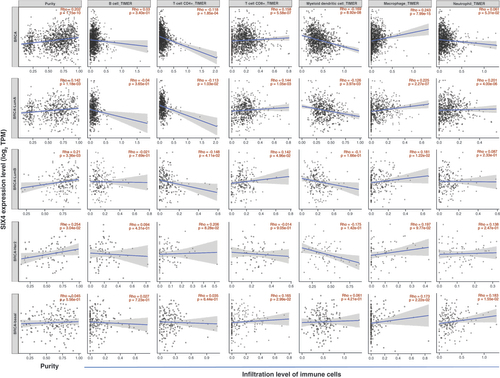
Expression of SIX4 negatively correlates with immune infiltration in breast cancer
The relationship of SIX4 with immune response in breast cancer was investigated by analyzing different types of infiltrating immune cells. Regardless of the molecular type of breast cancer, no correlation was found between infiltration of B cells and SIX4 expression level (). However, the infiltration of CD4+ T cells (Rho = -0.118; p = 1.85e-04) and myeloid dendritic cells (Rho = -0.169; p = 8.92e-08) was dramatically decreased in breast cancer patients with high SIX4 levels, while CD8+ T cells (Rho = 0.158; p = 5.58e-07) and macrophages (Rho = 0.243; p = 7.99e-15) increased with higher SIX4 levels.
Further analysis of different subtypes showed that infiltration of CD4+ T cells was suppressed in luminal A and luminal B subtype breast cancers (Rho = -0.113; p = 1.03e-02; and Rho = -0.148; p = 4.11e-02), while no significance was found in HER2+ and basal subtypes (p > 0.05). Conversely, infiltration of CD8+ T cells was increased in luminal A and luminal B breast cancer (Rho = 0.144; p = 1.05e-03; and Rho = 0.142; p = 4.96e-02), as well as in the basal subtype (Rho = 0.165; p = 2.99e-02), while no significance was found in HER2+ tumors (p > 0.05).
For myeloid dendritic cells, decreased infiltration was only found in the luminal A subtype (Rho = -0.126; p = 3.97e-03). The infiltrating pattern of macrophages was similar to that of CD8+ T cells, being increased in luminal A/B and basal subtypes (Rho >0, p < 0.05), and increased neutrophil infiltration was detected in luminal A and basal subtypes (Rho = 0.201; p = 4.00e-06; and Rho = 0.183; p = 1.55e-02).
High SIX4 protein level is associated with advanced progression of breast cancer
A total of 137 patients were recruited for further analysis after being evaluated by immunohistochemistry (A & ), with 48 in the low-expression group and 89 in the high-expression group (). There was no significant difference in expression of SIX4 among different age groups, ER/PR/HER2 statuses or molecular subtypes. Similar to the findings for expression of SIX4 mRNA, high protein levels of SIX4 tended to occur in patients with advanced disease, along with large tumor size and positive lymph node metastasis. Regarding breast cancer American Joint Committee on Cancer stage, expression of SIX4 was high in stage 3, compared with stages <3 (p = 0.0293).
(A & B) Representative staining of SIX4 in breast cancer tissues with (A) low and (B) high expression. (C) Protein–protein interactions of SIX4. (D–F) Overall survival of (D) all breast cancer patients, (E) ER− patients and (F) ER+ patients. (G) Staining of SIX4 in the Human Protein Atlas (HPA031794). (H) The protein expression of SIX4 in different malignancies.
ER: Estrogen receptor.
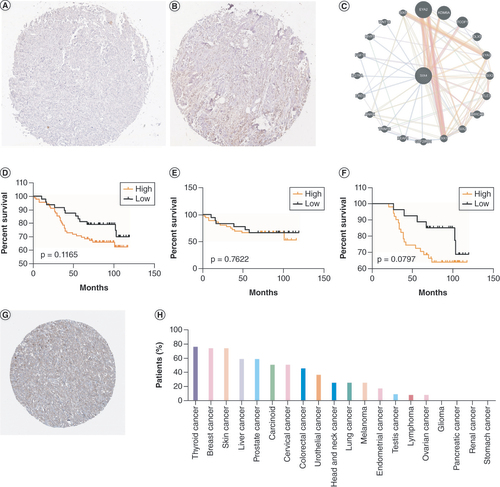
Table 1. The relationship between SIX4 protein level and clinicopathological parameters of breast cancer patients.
High protein levels of SIX4 suggest poor survival of breast cancer patients, especially ER+ patients
Based on PPI results, 20 proteins and 112 links were found to be potentially associated with SIX4; the 20 proteins were EYA2, KDM6A, TCOF1, DLX5, EYA3, SIX3, TLE3, LRRC46, SIX2, SIX1, GTF21RD1, KEAP1, TRIM16, ANHX, EREG, IL-27RA, FGF11, MYF5, WNT16 and SIX5 (C). Interestingly, other SIX family members were found in the PPI network with SIX4, indicating potential cooperation with SIX4 in regulating the progression of breast cancer. With the survival information of the recruited patients, to determine the prognostic value of SIX4, Kaplan–Meier survival curves were analyzed based on the levels of SIX4 in all patients, and separately for ER− and ER+ patients (D–F). Interestingly, although no statistical significance was found in any of the groups, a trend could be observed where high protein levels of SIX4 suggested poor survival of breast cancer patients (D), especially ER+ patients (F), which was similar to the prognostic value of SIX4 mRNA in breast cancer patients. The Human Protein Atlas provided the protein level of SIX4 in different types of malignancies, revealing the high expression of SIX4 in breast cancer (G), with breast cancer having the second highest expression (H).
Discussion
Immune escape, involving multiple mechanisms, concerns the inherent ability of tumor cells to hide from antitumor immune responses [Citation26]. As an immunogenic tumor, breast cancer has been classified to be immune responsive [Citation27]. Importantly, although immunotherapy has emerged as a promising therapeutic strategy for breast cancer patients, the benefits have been non-ideal, based on the results of current clinical trials [Citation26]; this may be due to the diverse characteristics of breast cancer subtypes.
Previous investigations have revealed that the genes involved in the differentiation of embryonic stem cells and organogenesis can regulate oncogenesis and development of malignancies [Citation28]. Fang et al. conducted a comprehensive analysis of SIX family members in CRC and proposed SIX4 as a potential therapeutic target for the treatment of CRC patients [Citation15]. Although the authors did not focus on breast cancer, a significant increase of SIX4 in breast cancer tissues was found, implicating its possible involvement in the oncogenesis and development of breast cancer [Citation15]. To investigate the involvement of SIX4 in breast cancer, the current study used GEPIA2 to verify the expression pattern of SIX4 in different types of malignancies. As expected, the expression of SIX4 was elevated in breast cancer tissues compared with normal tissue.
Based on these promising results, further investigation was conducted in different subgroups of breast cancer patients. Patients with advanced-stage disease tended to express high levels of SIX4, suggesting an oncogenic role of SIX4 in breast cancer. Camolotto et al. focused on pancreatic ductal adenocarcinoma (PDAC) and specified the role of HNF4-α, SIX4 and SIX1 in regulating the growth and molecular subtype of PDAC [Citation29]. Interestingly, they found that HNF4-α served as a tumor suppressor through repressing SIX4 and SIX1, two mesodermal/neuronal lineage specifiers expressed in the basal-like subtype of PDAC [Citation29], predicting a distinct function of SIX4 based on different subtypes of malignancies. For breast cancer, molecular subtypes based on the expression of hormone receptors is well accepted. In the present study, among different molecular subtypes, luminal breast cancers have the highest level of SIX4. In addition, in luminal breast cancer subtypes, high expression of SIX4 predicts poor survival. Genetic alteration analysis revealed that the methylation level of the SIX4 gene promoter was significantly decreased in breast cancer tissues. It is well known that DNA methylation is a crucial epigenetic modification to regulate the functioning of the genome [Citation30]. In the current investigation, we found that a low SIX4 gene promoter methylation level is negatively associated with its expression, especially in ER+ subtypes, indicating that increased SIX4 expression has an oncogenic role based on its decreased promoter methylation in different subtypes of breast cancer.
Allison et al. demonstrated that metabolic reprogramming in the tumor microenvironment (TME) is a hallmark of cancer cells as well as T lymphocytes [Citation31]. In patients with breast cancer, infiltration of CD4+ T cells is decreased according to increased SIX4 levels, especially in the luminal subtypes, consistent with the expression pattern and prognostic value of SIX4. CD4+ T cells belong to the category of T helper cells and are very important indicators that directly reflect the body’s immune status [Citation32]. As inflammatory CD4+ T cells can drive breast cancer into senescence through releasing IFN-γ and TNF-α, as well as directly binding to cancer cells, Boieri et al. revealed a novel mechanism to activate CD4+ T cell immunity against advanced breast cancer, showing promise for using the antitumor effect of CD4+ T cells for breast cancer immunotherapy [Citation33]. However, high levels of SIX4 are associated with reduced tumor infiltration of CD4+ T cells.
In contrast, the infiltration of CD8+ T cells increases with increasing SIX4 levels. Recently, Van der Leun et al. reviewed the state of CD8+ T cells in human cancer and proposed the dysfunction of T cells in cancer as being associated with a change in T-cell functionality rather than inactivity [Citation34]. Although exhaustion of CD8+ T cells has been observed in various types of malignancies [Citation35,Citation36], the underlying differentiation processes of cytotoxic cell states are not strictly tumor-specific or TME-dependent, and further investigation is required to clarify whether cytotoxic effector cells originate from a distinct pool of cells [Citation34]. In the current study, though increased, the CD8+ T cells might be dysfunctional, although more research is needed.
Myeloid dendritic cells, the most potent antigen-presenting cells in the body, decrease along with increases in SIX4 level. Dendritic cells are important in connecting innate to adaptive immune responses via the activation of T cells [Citation37]. The decreased infiltration of myeloid dendritic cells reduces the activation of T cells based on suppressing antigen presentation. On the other hand, the M2 phenotype of macrophages has the potential to promote the development of malignancy through enhancing the formation of an immunosuppressive myeloid microenvironment [Citation38]. The increased infiltration of macrophages along with SIX4 expression needs further investigation to distinguish the macrophage phenotypes. Based on the findings mentioned above, immunosuppressive conditions are likely formed with high SIX4 levels in breast cancer, and have the potential to promote the development and metastasis of tumor cells.
As a transcription factor, SIX4 is involved in organogenesis associated with the stemness of embryonic stem cells [Citation6]. Genes correlating with SIX4 were also related to stemness, which is also a key hallmark of malignancy. For regulation of the TME, stemness is also important for reducing antigen presentation and immune evasion [Citation39], indicating that increased SIX4 provides a prospective target for cancer therapy involving immune infiltration.
Conclusion
The transcription factor SIX4 is increased in patients with breast cancer, especially in the luminal subtypes. This upregulation is associated with low promoter methylation and correlates with immune infiltration. The stemness of SIX4 and SIX4-correlated genes identifies potential mechanisms of oncogenesis and development of breast cancer, providing a novel direction to prevent immune evasion and improve the immune response for novel immunotherapy. SIX4 may be involved in crosstalk between cancer stem cells and immune cells, resulting in an immunosuppressive microenvironment and providing a novel therapeutic target, particularly for luminal breast cancer patients, with immunotherapeutic intervention for controlled modulation of the immune system.
Human SIX4, a member of the homeobox family, functions as a transcription factor to regulate the differentiation or maturation of neuronal cells.
SIX4 expression is high in breast cancer tissues compared with normal tissues.
High SIX4 expression is associated with increased disease stage of breast cancer.
High levels of SIX4 predict poor survival of patients with breast cancer, especially for luminal subtypes.
SIX4 gene promoter methylation is negatively related to its expression in breast cancer.
SIX4 is enriched in transcriptional processes and immune response in breast cancer.
Expression of SIX4 negatively correlates with immune infiltration in breast cancer.
SIX4 may be involved in crosstalk between cancer stem cells and immune cells, resulting in an immunosuppressive microenvironment and providing a novel therapeutic target, particularly for luminal breast cancer patients.
Author contributions
H Wu, Y Cui and J Liu contributed to the conception and design of the study. H Wu conducted most of the experiments and analyzed the data. Z Wu, Z Fang, Y Hou, B Wu and Y Deng performed some of the experiments and interpreted the data. J Liu and Y Cui assisted with the experimental design and data analysis. H Wu and J Liu interpreted the data and drafted and revised the manuscript carefully. Y Cui and J Liu critically revised the original manuscript. All authors contributed to manuscript revision and read and approved the submitted version.
Writing disclosure
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations.
Acknowledgments
The authors are thankful to S Lin for his critical and careful editing and proofreading of the manuscript.
Financial disclosure
This work was supported by the National Natural Science Foundation of China (nos. 82273457 and 81501539), the Natural Science Foundation of Guangdong Province (nos. 2021A1515012180 and 2023A1515012762), Science and Technology Special Project of Guangdong Province (no. 210715216902829), Special Grant for Key Area Programs of Guangdong Education Department (no. 2021ZDZX2040) and ‘Dengfeng Project’ for the construction of high-level hospitals in Guangdong Province – First Affiliated Hospital of Shantou University College Supporting Funding (no. 202003-10). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Competing interests disclosure
The authors have no competing interests or relevant affiliations with any organization or entity with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
References
- Hanahan D , WeinbergRA. Hallmarks of cancer: the next generation. Cell144(5), 646–674 (2011).
- Siegel RL , MillerKD , FuchsHE , JemalA. Cancer statistics, 2022. CA Cancer J. Clin.72(1), 7–33 (2022).
- Onkar SS , CarletonNM , LucasPCet al. The great immune escape: understanding the divergent immune response in breast cancer subtypes. Cancer Discov.13(1), 23–40 (2022).
- Wurmser M , ChaverotN , MadaniRet al. SIX1 and SIX4 homeoproteins regulate PAX7+ progenitor cell properties during fetal epaxial myogenesis. Development147(19), dev185975 (2020).
- Chen R , HouY , ConnellM , ZhuS. Homeodomain protein Six4 prevents the generation of supernumerary Drosophila type II neuroblasts and premature differentiation of intermediate neural progenitors. PLoS Genet.17(2), e1009371 (2021).
- Magli A , BaikJ , MillsLJet al. Time-dependent Pax3-mediated chromatin remodeling and cooperation with Six4 and Tead2 specify the skeletal myogenic lineage in developing mesoderm. PLoS Biol.17(2), e3000153 (2019).
- Takahashi M , TamuraM , SatoS , KawakamiK. Mice doubly deficient in Six4 and Six5 show ventral body wall defects reproducing human omphalocele. Dis. Model. Mech.11(10), dmm034611 (2018).
- Liu P , CaiS , LiN. Circular RNA-hsa-circ-0000670 promotes gastric cancer progression through the microRNA-384/SIX4 axis. Exp. Cell Res.394(2), 112141 (2020).
- Han J , HuX. IGF2BP3-stabilized SIX4 promotes the proliferation, migration, invasion and tube formation of ovarian cancer cells. Mol. Med. Rep.26(1), 232 (2022).
- Wang Z , SunBS , ChenZSet al. FOXA1 leads to aberrant expression of SIX4 affecting cervical cancer cell growth and chemoresistance. Anal. Cell. Pathol. (Amst.)2022, 9675466 (2022).
- He Q , LinZ , WangZet al. SIX4 promotes hepatocellular carcinoma metastasis through upregulating YAP1 and c-MET. Oncogene39(50), 7279–7295 (2020).
- Tang X , YangY , SongXet al. SIX4 acts as a master regulator of oncogenes that promotes tumorigenesis in non-small-cell lung cancer cells. Biochem. Biophys. Res. Commun.516(3), 851–857 (2019).
- Sun XL , MaJ , ChenQZet al. SIX4 promotes metastasis through STAT3 activation in breast cancer. Am. J. Cancer Res.10(1), 224–236 (2020).
- Li Y , JiangX , YanX , WangY. Upregulation of SIX4 indicates poor clinical outcome and promotes tumor growth and cell metastasis in esophageal squamous cell carcinoma. Thorac. Cancer12(6), 752–759 (2021).
- Fang ZX , LiCL , WuZ , HouYY , WuHT , LiuJ. Comprehensive analysis of the potential role and prognostic value of sine oculis homeobox homolog family in colorectal cancer. World J. Gastrointest. Oncol.14(11), 2138–2156 (2022).
- Cao Y , ChenC , TaoY , LinW , WangP. Immunotherapy for triple-negative breast cancer. Pharmaceutics13(12), 2003 (2021).
- Karlsson M , ZhangC , MearLet al. A single-cell type transcriptomics map of human tissues. Sci. Adv.7(31), eabh2169 (2021).
- Chandrashekar DS , KarthikeyanSK , KorlaPKet al. UALCAN: an update to the integrated cancer data analysis platform. Neoplasia25, 18–27 (2022).
- Lanczky A , GyorffyB. Web-based survival analysis tool tailored for medical research (KMplot): development and implementation. J. Med. Internet Res.23(7), e27633 (2021).
- Asleh K , LluchA , GoytainAet al. Triple negative PAM50 non-basal breast cancer subtype predicts benefit from extended adjuvant capecitabine. Clin. Cancer Res.29(2), 389–400 (2022).
- Cerami E , GaoJ , DogrusozUet al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov.2(5), 401–404 (2012).
- Vasaikar SV , StraubP , WangJ , ZhangB. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res.46(D1), D956–D963 (2018).
- Warde-Farley D , DonaldsonSL , ComesOet al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res.38(Web Server issue), W214–220 (2010).
- Li T , FuJ , ZengZet al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res.48(W1), W509–W514 (2020).
- Liu J , WeiXL , HuangWH , ChenCF , BaiJW , ZhangGJ. Cytoplasmic Skp2 expression is associated with p-Akt1 and predicts poor prognosis in human breast carcinomas. PLOS ONE7(12), e52675 (2012).
- Chen DS , MellmanI. Elements of cancer immunity and the cancer-immune set point. Nature541(7637), 321–330 (2017).
- Miller LD , ChouJA , BlackMAet al. Immunogenic subtypes of breast cancer delineated by gene classifiers of immune responsiveness. Cancer Immunol. Res.4(7), 600–610 (2016).
- Gerrard DT , BerryAA , JenningsREet al. Dynamic changes in the epigenomic landscape regulate human organogenesis and link to developmental disorders. Nature Commun.11(1), 3920 (2020).
- Camolotto SA , BelovaVK , Torre-HealyLet al. Reciprocal regulation of pancreatic ductal adenocarcinoma growth and molecular subtype by HNF4alpha and SIX1/4. Gut70(5), 900–914 (2021).
- Ehrlich M . DNA hypermethylation in disease: mechanisms and clinical relevance. Epigenetics14(12), 1141–1163 (2019).
- Allison KE , CoomberBL , BridleBW. Metabolic reprogramming in the tumour microenvironment: a hallmark shared by cancer cells and T lymphocytes. Immunology152(2), 175–184 (2017).
- Mamula D , KhosousiS , HeY , LazarevicV , SvenningssonP. Impaired migratory phenotype of CD4+ T cells in Parkinson’s disease. NPJ Parkinsons Dis.8(1), 171 (2022).
- Boieri M , MarcheseE , PhamQMet al. Thymic stromal lymphopoietin-stimulated CD4+ T cells induce senescence in advanced breast cancer. Front. Cell Dev. Biol.10, 1002692 (2022).
- Van der Leun AM , ThommenDS , SchumacherTN. CD8+ T cell states in human cancer: insights from single-cell analysis. Nat. Rev. Cancer20(4), 218–232 (2020).
- Baitsch L , BaumgaertnerP , DevevreEet al. Exhaustion of tumor-specific CD8+ T cells in metastases from melanoma patients. J. Clin. Invest.121(6), 2350–2360 (2011).
- Blackburn SD , ShinH , FreemanGJ , WherryEJ. Selective expansion of a subset of exhausted CD8 T cells by alphaPD-L1 blockade. Proc. Natl Acad. Sci. USA105(39), 15016–15021 (2008).
- Lee M , DuH , WinerDA , Clemente-CasaresX , TsaiS. Mechanosensing in macrophages and dendritic cells in steady-state and disease. Front. Cell Dev. Biol.10, 1044729 (2022).
- Liu M , LiuL , SongY , LiW , XuL. Targeting macrophages: a novel treatment strategy in solid tumors. J. Transl. Med.20(1), 586 (2022).
- Dianat-Moghadam H , MahariA , SalahlouR , KhaliliM , AziziM , SadeghzadehH. Immune evader cancer stem cells direct the perspective approaches to cancer immunotherapy. Stem Cell Res. Ther.13(1), 150 (2022).