Abstract
Mammalian DNA duplexes are highly condensed with different components, including histones, enabling chromatin formation. Chromatin remodeling is involved in multiple biological processes, including gene transcription regulation and DNA damage repair. Recent research has highlighted the significant involvement of really interesting new gene (RING) finger proteins in chromatin remodeling, primarily attributed to their E3 ubiquitin ligase activities. In this review, we highlight the pivotal role of RING finger proteins in chromatin remodeling and provide an overview of their capacity to ubiquitinate specific histones, modulate ATP-dependent chromatin remodeling complexes and interact with various histone post-translational modifications. We also discuss the diverse biological effects of RING finger protein-mediated chromatin remodeling and explore potential therapeutic strategies for targeting these proteins.
Chromatin, the fundamental structure of eukaryotic cells, consists of DNA, histones and nonhistone proteins, and plays a crucial role in maintaining genetic stability, regulating gene expression and transmitting genetic information. Chromatin is classified into euchromatin and heterochromatin. The loosely arranged chromatin structure of euchromatin allows transcription factors and regulators easy access to DNA sequences, leading to enhanced transcription and gene expression. Conversely, heterochromatin is a condensed state of chromatin that inhibits gene transcription by limiting the access of transcription factors to DNA binding sites. Chromatin remodeling modulates gene expression by regulating the dynamic transition process between euchromatin and heterochromatin at specific developmental stages or cell states [Citation1].
Nucleosomes occupy a central role in the organization of chromatin. The dynamic substance of chromatin packages and regulates the genetic material within the cell nucleus. At the core of the nucleosome, the fundamental structural unit consists of an octamer composed of four pairs of histones: histone H2A, histone H2B, histone H3 and histone H4. This octamer acts as a spool around which DNA is wrapped, creating a condensed chromatin structure. The interplay of nucleosomes and their associated proteins governs essential cellular processes by modulating access to the underlying DNA. The functional flexibility of chromatin is evident through numerous modifications that can be applied to its components. These modifications orchestrate a symphony of molecular events that facilitate chromatin remodeling and precisely regulate gene expression. These processes are mainly affected by DNA methylation, post-translational modifications (PTMs) of histone and nonhistone proteins, ATP-dependent chromatin remodeling complexes, and crosstalk among different types of histone modifications.
Histone PTMs play a key role in the regulation of chromatin. Histones undergo a wide array of PTMs, including methylation, acetylation, ubiquitination and phosphorylation. These chemical modifications serve as dynamic switches, affecting chromatin compaction and gene accessibility. In addition, the crosstalk between various types of histone PTMs increases the complexity of the regulatory landscape, whereby one modification can affect the placement or removal of others [Citation2,Citation3]. For instance, histone ubiquitination, the addition of ubiquitin molecules to histones, plays a crucial role in gene transcriptional regulation, DNA damage repair and the overall maintenance of chromatin integrity [Citation4]. It acts as a signal for various cellular processes, highlighting the complex network of interactions within the chromatin environment. Thus, nucleosomes, with their histone octamer cores, constitute the fundamental building blocks of chromatin, and the intricate modifications applied to these components finely orchestrate gene regulation. Understanding the language of nucleosome modifications is crucial to unravel the complexity of cellular processes, ranging from gene expression to DNA repair.
Really interesting new gene (RING) finger proteins are the most commonly found E3 ubiquitin ligases that are characterized by the RING finger domain. In addition to promoting ubiquitin transfer from the ubiquitin-conjugating enzyme E2 to the target protein, RING finger proteins regulate protein degradation and participate in cellular processes such as cell cycle progression, signal transduction and DNA repair. RING E3 ubiquitin ligases can be categorized into different subtypes depending on their mechanisms of action and associated protein domains. RING finger protein E3 ligases, including NEDD4 [Citation5], MDM2 [Citation6] and TRIM proteins [Citation7], independently and directly interact with target proteins. RING finger protein E3 ligase-containing complexes provide substrate recognition and binding functions, including in complexes such as APC/C and SCF. By elucidating the role of RING E3 ligases, we can achieve a better understanding of their biological functions and design more effective therapeutic interventions for cancers and autoimmune diseases [Citation8].
In this review, we summarize the modulation of RING finger proteins in chromatin remodeling by ubiquitinating different histones, regulating ATP-dependent chromatin remodeling complexes and engaging with other histone PTMs. Furthermoew, we discuss the diverse biological effects caused by RING finger protein-regulated chromatin remodeling and potential therapeutic approaches to RING finger protein targeting.
RING finger proteins ubiquitinate histones as E3 ubiquitin ligases
RING finger proteins mediate histone ubiquitination through their ubiquitin ligase activity, modulating chromatin structure and regulating diverse biological processes. Histone ubiquitination occurs primarily at specific histones, which has distinct implications for gene regulation [Citation9,Citation10], DNA replication [Citation11], DNA damage repair [Citation12,Citation13] and chromatin structure [Citation14–16]. We have summarized RING finger proteins ubiquitinating different lysine (Lys) sites of core histones (&).
As E3 ubiquitin ligases, RING finger proteins ubiquitinate different Lys (K) sites of histones. The RING finger proteins corresponding to the ubiquitinated histone Lys sites are shown. Also see also for references.
K: Lysine; Ub: Ubiquitin; Unknown: Lysine site remains unknown.
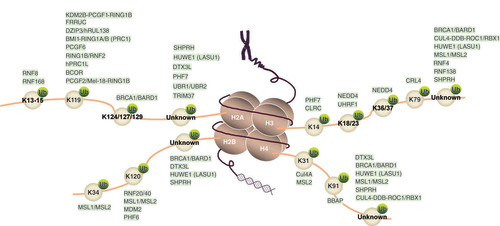
Table 1. RING finger proteins ubiquitinate different lysine sites of core histones.
Regulation of histone H2A ubiquitination by RING finger proteins
Histone H2A ubiquitination contributes to gene silencing or repression. One well-known form of histone H2A ubiquitination is the monoubiquitination of Lys-119 (H2AK119Ub). H2AK119Ub promotes chromatin compaction and recruits additional chromatin modifiers as a transcriptionally repressive mark for gene repression [Citation24]. RING domain-containing E3 ubiquitin ligases are the primary catalytic enzymes for histone H2A ubiquitination. RING1B (RING2, encoded by RNF2) is essential to histone modification and gene regulation by facilitating H2AK119Ub mediation [Citation18,Citation24,Citation58–61]. PRC1, an E3 core subunit of RING1A/B, monoubiquitinates histone H2AK119 by forming a complex with the E2 enzyme UbcH5c [Citation62,Citation63], which is involved in chromatin inactivation [Citation17] and silencing of HOX family genes [Citation18,Citation24,Citation64]. In addition, hPRC1L, containing RING1A and RING1B, monoubiquitinates nucleosomal histone H2AK119, resulting in transcriptional repression of drosophila Ubx [Citation18]. BCOR, consisting of RING1B, mediates histone H2AK119 monoubiquitination, further inhibiting specific transcription targets such as HOXA cluster genes, inducing heritable changes in chromatin [Citation65].
In addition, the PCGF2–RING1B complex, composed of two ubiquitin E3 ligases, RING1B and PCGF2 (also known as RNF110), significantly enhances ubiquitin ligase activity through nonspecifically ubiquitylating free histone substrates and highly specifically targeting histone H2A Lys-119 within nucleosomes. Mutation analysis indicates that RING1B is necessary to perform the E3 function, but PCGF2 directs this activity toward histone H2AK119 on chromatin [Citation20]. The RING finger protein 2A-HUB (also known as RING-type E3 ubiquitin transferase DZIP3) catalyzes histone H2AK119 monoubiquitination [Citation66], further repressing a subset of chemokine gene expression.
In addition to Lys-119, histone H2A ubiquitination can occur at other Lys residues. For example, BRCA1 facilitates the formation of polyubiquitin chains linked by Lys-6. The E3 ligase activity of the BRCA1–BARD1 complex is significantly enhanced as a result of the heterodimerization between BRCA1 and BARD1 [Citation67]. BRCA1–BARD1 ubiquitinates histone H2A specifically at Lys residues 127–129 in vitro and in vivo, causing further silencing at DNA satellite repeat regions by accumulating ubiquitin conjugate complex at DNA damage sites [Citation28]. RNF8 and RNF168 modulate chromatin structure by ubiquitinating histone H2A at Lys-13/Lys-15 [Citation26,Citation68]. TRIM37 is a novel histone H2A ubiquitin ligase that mediates the binding of PRC2 to specific target genes, such as Fas [Citation7], with unknown specific Lys sites. Furthermore, the RING-type E3 ligases UBR1 and UBR2 facilitate either the monoubiquitination or polyubiquitination of histone H2A in vitro [Citation38], contributing to transcriptional repression during cell meiosis and chromatin inactivation [Citation39], although the precise Lys site at which UBR2 ubiquitinates histone H2A remains unknown.
Regulation of histone H2B ubiquitination by RING finger proteins
RING finger proteins ubiquitinate histone H2B, leading to changes in chromatin dynamics and gene expression. Histone H2B ubiquitination occurs mainly at Lys-120 (Lys-123 for budding yeast), which promotes transcriptional activation by inhibiting chromatin compaction and promoting the formation of transcriptionally active chromatin [Citation69,Citation70]. RNF40 and its paralog RNF20 catalyse histone H2B monoubiquitination at Lys-120 by the E2 enzyme UbcH6, resulting in the formation of H2BK120Ub (also known as H2Bub1) and promoting HOX gene expression [Citation44]. The RING finger E3 ubiquitin ligase MDM2 also possesses histone H2BK120 ubiquitination activity. MDM2 directly monoubiquitinates histone H2B by interacting with p53 [Citation6], exerting transcriptional repression via its RING domain [Citation46]. Furthermore, acting as a RING finger protein, MSL2 can form a complex with MSL1 that mediates ubiquitination at Lys-34 of histone H2B, resulting in the transcriptional activation of HOXA9 and MEIS1 [Citation47].
RING finger proteins mediate the ubiquitination of histones H3&H4
The ubiquitination of histone H3 and histone H4 by RING finger proteins impacts multiple cellular processes, such as transcriptional activation, gene stability maintenance, transcriptional inactivation, DNA replication and DNA damage repair. The E3 ubiquitin protein ligase NEDD4 mediates ubiquitination on histone H3 at the Lys-23/36/37 residues [Citation5] depending on its homologous to the E6-AP C-terminus (HECT domain). Through its DCAF8 (DDB1 and CUL4-associated factor 8) substrate receptor, the RING finger protein CRL4 performs ubiquitination on histone H3K79 within hepatocytes, resulting in the inactivation of fetal liver genes [Citation52]. UHRF1 exerts ubiquitin ligase activity via its RING domain and ubiquitinates histone H3 at Lys-23 [Citation11] and Lys-18 [Citation51] in mammalian cells.
RING finger proteins can also ubiquitinate histone H4. The complex comprising CUL4, DDB and ROC1 mediates ubiquitination on histone H3 and histone H4, resulting in the release of these proteins from nucleosomes, further impairing the interaction between histones and DNA, and allowing repair proteins to reach damaged foci [Citation16]. Furthermore, monoubiquitination of histone H4 at Lys-31 inhibits Mg2+-dependent chromatin compaction, similar to the suppression of chromatin compaction by H2BK120Ub [Citation71].
Thus, RING finger proteins play a significant role in the ubiquitination of core histones that regulate chromatin remodeling. Multifaceted processes of histone ubiquitination impact gene expression, DNA repair, chromatin organization and other cellular processes, emphasizing the importance of regulating histone ubiquitination to maintain proper cellular function.
RING finger proteins regulate ATP-dependent chromatin remodeling complexes
RING finger proteins are involved in regulating several chromatin-remodeling complexes, such as SWI/SNF and FACT.
The SWI–SNF complex facilitates the transcription of a specific group of genes that depend on RNF20 and H2BK120Ub at the transcriptional level [Citation72]. SMARCAD1 is activated in this context. The proximal coupling of ubiquitin conjugation to ER degradation (CUE) domain of SMARCAD1 recognizes the monoubiquitination of histone H2AK127/129 [Citation73], depending on its ATPase activity to not only cleave DNA breaks but also promote DNA repair with high fidelity [Citation74]. Moreover, the newly identified RING finger protein UNK ubiquitinates the SWI/SNF protein BAF60b in the presence of Rac GTPase [Citation75]. This interaction highlights how RING finger proteins can directly influence the activities of chromatin-remodeling complexes, ultimately reshaping chromatin structure.
In a chromatin-mediated transcription system, the monoubiquitination of histone H2B by RNF20/40 is essential for regulating the transcription elongation factor PAF and the histone chaperone FACT [Citation9]. Histone H2B monoubiquitination enables FACT to stimulate transcriptional elongation [Citation9]. Beyond its involvement in transcriptional regulation, RNF20 remodels chromatin structure to permit homologous recombination (HR) repair proteins to assess DNA for repair in eukaryotes by ubiquitinating histone H2B [Citation10]. Notably, a key component of FACT, SUPT16H, directly binds to RNF20. This interaction further underscores the close relationship between RING finger proteins and chromatin-remodeling complexes, with SUPT16H contributing to chromatin remodeling specifically for HR repair [Citation76].
Therefore, the complex interaction between RING finger proteins and chromatin-remodeling complexes has significant implications for regulating gene transcription, repairing DNA damage and shaping chromatin structure.
Crosstalk between RING finger protein-mediated histone ubiquitination&other histone PTMs
Histone PTMs may interact and lead to different outcomes. This section will discuss the crosstalk between histone ubiquitination mediated by RING finger proteins and other histone PTMs, such as histone methylation and histone acetylation (,&&).
Different K sites of histones can be chemically modified by adding different small molecules. Ub and the corresponding RING finger proteins are shown in green, Me and the corresponding methyltransferases in orange, and Ac and the corresponding acetyltransferases in blue. Sharp arrows indicate facilitation; flat arrows indicate inhibition.
Ac: Acetyl; K: Lysine; Me: Methyl; Ub: Ubiquitination.
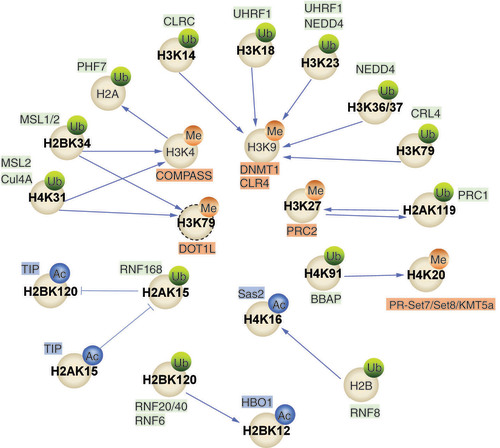
Table 2. RING finger proteins mediate the crosstalk between histone ubiquitination and histone methylation; the relationship between the interactions is also shown.
Table 3. RING finger proteins that mediate the crosstalk between histone ubiquitination and histone acetylation; the relationship between the interactions is also listed.
Crosstalk between RING finger protein-mediated histone ubiquitination&histone methylation
The complex interaction between histone ubiquitination mediated by RING finger proteins and the process of histone methylation is a noteworthy and crucial aspect of chromatin regulation. These two distinct PTMs work together to affect chromatin structure and gene expression by orchestrating a complex molecular conversation. Histone ubiquitination, catalyzed by RING finger E3 ligases, involves attaching ubiquitin moieties covalently to specific Lys residues on histones. This modification can lead to various outcomes, such as changes in nucleosome stability, the recruitment of effector proteins and DNA repair processes. In parallel, histone methylation, governed by histone methyltransferases, introduces methyl groups to specific histone Lys or arginine residues. Histone methylation signifies chromatin regions for distinct functional roles, influencing gene transcription and other chromatin-associated activities. Interestingly, these two modifications are not isolated events but are intricately linked through various mechanisms (&).
Polycomb group (PcG) proteins are a group of transcription repressors that regulate target genes through chromatin modification. PcG proteins can be grouped into two major core protein complexes, PRC1 and PRC2, based on their biochemical and functional properties. These complexes play a crucial role in gene silencing through epigenetic processes and are integral to the regulation of gene expression in eukaryotic organisms. PRC1 is a macromolecular assembly consisting of multiple protein subunits, each with distinct roles in initiating gene silencing. One of the primary roles of this enzyme is facilitating the catalysis of monoubiquitination on histone H2A at Lys-119 (H2AK119Ub) [Citation17]. By introducing this ubiquitin mark, PRC1 reinforces chromatin compaction and hinders the access of transcriptional machinery to target genes. PRC2 enhances the gene-silencing capabilities of PRC1 through its catalytic function in the di- and trimethylation of histone H3 at lysine 27 (H3K27me2/3) [Citation81,Citation82]. It is well documented that H2AK119Ub promotes recruiting PRC2-mediated H3K27me3 [Citation77,Citation78], while PRC2-mediated H3K27me3 facilitates PRC1-mediated H2AK119Ub [Citation81,Citation82]. PHF7 is a recently discovered E3 ligase that can selectively ubiquitinate histone H2A by binding to H3K4me3/me2 before the transition from histones to protamine occurs [Citation40].
RNF20/40-catalyzed H2BK120Ub facilitates the recruitment of the histone methyltransferase COMPASS to achieve H3K4me2 and H3K4me3 [Citation50,Citation83,Citation84], which forms a network of crosstalk between different histone modifications that contributes to the regulation of gene expression. For instance, MSL2-mediated H2BK34Ub [Citation51] and Cul4A-mediated H4K31Ub [Citation74] promote histone H3K4 and histone H3K79 methylation during histone crosstalk. Notably, histone H3K79 is one of the histone core sites that can be methylated, and this modification is catalyzed by Dot1L [Citation85]. Importantly, the catalytic activity of Dot1L is dependent on H2BK120Ub, providing a fascinating example of crosstalk between histone PTMs [Citation86,Citation87]. Recently, significant progress has been made in understanding the structural basis of crosstalk between H2BK120Ub and methylation by depicting the cryo-electron microscopy structure of Dot1L in nucleosomes containing H2BK120Ub in both its active and inactive states. These findings provide insight into the mechanisms underlying the enhancement of histone H3K79 methylation by H2BK120Ub [Citation88–91]. Specifically, in the active state, Dot1L establishes specific interactions with the ubiquitin moiety linked to histone H2BK120 and histone H3K79 [Citation88].
Moving beyond H2BK120Ub, UHRF1 mediates histone H3K18/23 ubiquitination due to its histone- and DNA-binding activities [Citation11]. Through the SRA domain [Citation92], UHRF1 engages DNMT1 at hemi-methylation sites [Citation93,Citation94], thereby maintaining DNA methylation [Citation11,Citation95]. In addition, UHRF1 has been identified as a novel H3K9m-specific binding protein that is responsible for the formation of heterochromatin in mammalian cells via its PHD and SRA domain [Citation96]. BBAP bestows a single ubiquitin molecule onto Lys-91 of histone H4. This action elicits a safeguarding reaction against DNA-damaging substances while fostering the association between 53BP1 and methylated histone H4K20 [Citation56]. Upon DNA damage, BARD1 interacts with Lys-9-dimethylated histone H3 (H3K9me2) via a serine-protein kinase ATM-dependent mechanism that is not affected by RNF168 [Citation97].
Crosstalk between RING finger protein-mediated histone ubiquitination&histone acetylation
Acetylation is a crucial PTM that regulates both chromatin structure and function. The dynamic interplay between RING finger protein-mediated histone ubiquitination and the complicated process of histone acetylation is pivotal for chromatin modulation ( & ). This interplay between these histone modifications represents a vital regulatory mechanism for chromatin dynamics and gene expression.
RNF168 mediates the ubiquitination of histone H2AK15 and inhibits histone H2BK120 acetylation, which is mediated by TIP60 [Citation79]. Coincidentally, TIP60 can also mediate the acetylation of histone H2AK15 (H2AK15Ac) in vivo, and in turn, H2AK15Ac negatively regulates histone H2AK15 ubiquitination [Citation79]. This intricate interplay highlights the reciprocal relationship between histone ubiquitination and acetylation. Histone H3 takes center stage in another fascinating interaction. By catalyzing ubiquitination on Lys residues 23/36/37 of histone H3, NEDD4 facilitates the recruitment of histone acetyltransferase GCN5, which leads to the subsequent acetylation of histone H3K9 and the transcriptional activation of some genes, including IL-1α, IL-1β and GCLM [Citation5]. Here, the interplay between the ubiquitination and acetylation of histone finely regulates gene expression, highlighting the complexity of chromatin regulation.
The TIP60–UBC13 complex ubiquitinates histone H2AX (depending on prior histone H2AX acetylation), releases histone H2AX from damaged chromatin and facilitates DDR [Citation98]. The precise coordination between these modifications emphasizes their functional importance in DNA-repair processes. Moreover, PHF6 introduces another level of complexity in recognizing H2BK12Ac, which in turn facilitates its E3 ubiquitin ligase function [Citation45]. The ubiquitination of histones controlled by RNF8 triggers histone H4K16 acetylation, potentially serving as the primary phase of nucleosome elimination [Citation80]. The E3 ligase UHRF2 upholds the stability of the acetyltransferase TIP60, regulating the levels of H3K9Ac and H3K14Ac through its RING finger domain [Citation99].
The crosstalk between histone ubiquitination and methylation or acetylation reflects a layer of complexity in the complex regulatory landscape of chromatin biology. The interaction between histone ubiquitination and other histone PTMs elicits a cascade activation or inhibitory effect through mutual interactions, promoting or weakening each other’s function, and ultimately affecting gene transcription. This crosstalk enables the integration of diverse signals and pathways, affording precise control over gene expression in response to alterations in cellular cues. Understanding the intricate mechanisms regulating chromatin dynamics and their impact on development, disease and cellular functions can provide valuable insights. Unfortunately, the exact regulatory mechanism of the crosstalk is still poorly understood, and more in-depth analysis and exploration are still needed.
Biological functions of RING finger proteins in regulating chromatin remodeling
RING finger protein-mediated histone ubiquitination regulates gene transcription
Chromatin is a complex of DNA and associated proteins, consisting mainly of histones, that condense the DNA into a highly organized structure. The degree of chromatin condensation is a key factor in determining whether a gene is available for transcription (gene activation) or not (gene repression). Under normal physiological conditions, histone ubiquitination contributes to the fine-tuned control of the efficient expression of genes required for normal cellular processes. Chromatin structure alterations play a crucial role in regulating gene expression at the molecular level. These alterations, including changes in nucleosome positioning, histone modifications, DNA methylation and chromatin looping, have direct impacts on the extent of chromatin condensation, which in turn leads to changes in gene expression. RING finger protein-mediated histone ubiquitination can alter the structure of chromatin. RING finger protein can promote either an open or closed chromatin structure, depending on ubiquitinating the specific Lys residue of ubiquitinated histones. Consequently, changes in chromatin structure can directly affect the accessibility of genes to transcriptional machinery.
H2AK119Ub serves as a specific marker for epigenetic transcriptional repression, and histone H2B ubiquitination is a marker for transcriptional activation [Citation100,Citation101]. Histone H2AK119 ubiquitination mediated by RING finger proteins, including RING2-containing complexes (PRC1, hPRC1L, BCOR), PCGF2-RING1B, and RING finger protein 2A-HUB, UBR1 and UBR2, is strongly associated with transcriptional repression. Mechanistically, RING2-containing complex-catalyzed histone H2AK119 ubiquitination inhibits the activated form of the RNA polymerase II preinitiation complex [Citation79] by blocking an early and integral event in preinitiation complex formation [Citation102,Citation103]. Histone H2AK119 ubiquitination enhances higher order chromatin compaction by promoting histone H1 binding with nucleosomes, thus further repressing gene transcription [Citation104]. Furthermore, ubiquitination of histone H2AK119 catalyzed by 2A-HUB represses transcription by preventing FACT recruitment to the promoter region, hindering the elongation of RNA polymerase II during transcription [Citation66].
The interaction between RNF20–RNF40 and RNA PAF1 together with RNA polymerase II at chromatin facilitates transcriptional elongation [Citation9]. In addition, FACT is directed to chromatin sites with H2BK120Ub modification, causing physical disruption of the histone H2A–H2B dimer, leading to the reconstruction of a more open state of chromatin, thereby facilitating the crossing of RNA polymerase II and promoting transcriptional activation [Citation9].
Therefore, the regulation of gene transcription is facilitated by RING finger protein-mediated histone ubiquitination, which interferes with the recruitment of transcription factors and RNA polymerase II and subsequently modifies chromatin remodeling.
RING finger protein-mediated histone ubiquitination participates in DDR
DDR signaling pathways are activated when cells are disturbed by environmental and endogenous factors. The incidence of DNA double-strand breaks [Citation105] results in genome instability [Citation106]. RING finger proteins are involved in DDR by interacting with DNA repair and HR proteins, coordinating structural changes in chromatin.
The ubiquitination of histone H2AK119, histone H2AK13/15 and histone H2AK127/129 plays critical functions in DDR. RING2–BMI1 is responsible for ubiquitinating histone H2AK119 and histone H2AX, consequently promoting double-strand break repair [Citation107,Citation108]. Histone H2A/H2AXK13/15 ubiquitination catalyzed by RNF8 and RNF168 is an essential signal in DDR. In a physiological context, RNF168 alters chromatin architecture through ubiquitinating histone H2A [Citation68]. However, when DNA damage occurs, RNF168 is mobilized to damaged DNA loci, increasing the abundance of ubiquitinated proteins and initiating the downstream signaling pathways [Citation109]. RNF8 first polyubiquitinates histone H1 (K63-linked ubiquitin chain) to recruit RNF168, monoubiquitinates histone H2A/H2AXK13/15 and finally extends the K63-linked ubiquitin chain by RNF8 to activate repair factors [Citation68,Citation110]. Furthermore, BRCA1 is associated with BARD1 [Citation52] and is located at the central region of the lesion with HP1α to catalyze histone H2AK127/129 monoubiquitination. Histone H2AK127/129 monoubiquitination by the CUE domain of SMARCAD1 [Citation73] cleaves DNA breaks and promotes DNA repair with high fidelity depending on its ATPase activity [Citation74].
H2BK120Ub is related to transcriptional activation and acts as a signal to actively recruit DNA damage-associated proteins [Citation111]. Histone H2BK120 monoubiquitination regulated by RNF20–RNF40 contributes to mammalian DDR [Citation10], which facilitates histone H3K79 dimethylation for recruiting BRCA1, 53BP1 and RAD51 to DNA end resection for strand invasion [Citation112–114]. RNF20-dependent histone H2B ubiquitination is also required for HR [Citation111,Citation115]. Moreover, the crosstalk between histone H2BK120 ubiquitination by RNF20–RNF40 and histone H3 methylation contributes to cell cycle arrest in response to DNA damage [Citation116]. CUL4–DDB–ROC1 abolishes the interaction between histones and DNA, ultimately facilitating the effective enlistment of repair proteins to areas of DNA damage [Citation16].
In yeast, Rad5 as an ATP-dependent chromatin remodeler [Citation117] possesses a RING finger domain that confers its E3 ubiquitin-protein ligase activity [Citation118]. Rad5 can polyubiquitinate PCNA and plays a critical role in postreplication repair [Citation119]. Two mammalian Rad5 homologs are SHPRH and HLTF [Citation120]. SHPRH and HLTF mediate polyubiquitination of PCNA at Lys 164 via activating a pathway of error-free template switching during DNA replication [Citation120–122]. SHPRH also participates in DNA repair and transcriptional regulation through chromatin ubiquitination [Citation123,Citation124].
Therefore, RING finger protein-mediated histone ubiquitination participates in DDR by providing a signal for DNA damage, recruiting repair factors to the damaged site and facilitating the repair process through chromatin remodeling. This involvement ensures the maintenance of genome integrity and prevents mutations and genomic instability, which may cause diseases such as cancer.
Development of targeted therapeutics for RING finger proteins
RING finger proteins are widely implicated in the onset and progression of multiple diseases, including cancer [Citation8], and thus have the potential to become therapeutic targets. The PRC1 protein BMI1 can ubiquitinate histone H2AK119 and its high expression is correlated with shorter survival duration in acute myeloid leukemia cells [Citation125]. A BMI1 inhibitor, unesbulin (also known as PTC596), has entered phase II clinical trials evaluating its effects against ovarian cancer (NCT03206645), leiomyosarcoma (NCT03761095), high-grade glioma and diffuse intrinsic pontine glioma (NCT03605550) [Citation126].
A recent study has identified UHRF1 as a novel epigenetic target for malignant pleural mesothelioma (MPM) [Citation127]. Studies have shown that mithramycin can reduce UHRF1 expression in vitro and in vivo, thus presenting a promising approach for targeting UHRF1 in MPM, although thus far no clinical trials evaluating UHRF1 have been conducted [Citation127]. Interestingly, circUHRF1 is found to be present in an exosomal manner in the plasma of patients with hepatocellular carcinoma (HCC) [Citation128]. HCC-derived exosomal circUHRF1 can induce natural killer (NK) cell exhaustion and decrease NK cell function and tumor infiltration, contributing to HCC resistance to anti-PD1 therapy [Citation128]. Mechanistically, circUHRF1 inhibits NK cell-derived IFN-γ and TNF-α secretion and drives resistance toanti-PD1 immunotherapy in HCC patients [Citation128]. Furthermore, circUHRF1 inhibits NK cell function by upregulating the expression of TIM-3 via the degradation of miR-449c-5p [Citation128]. Thus, circUHRF1 is associated with immunosuppression by decreasing NK cell proportion and tumor infiltration, further guiding the resistance to anti-PD1 immunotherapy in HCC patients [Citation128]. These emerging roles of UHRF1 and circUHRF1 in cancer, particularly in the context of MPM and HCC, offer exciting prospects for the development of novel therapeutic approaches [Citation128]. The potential of mithramycin as a UHRF1 inhibitor in MPM is a promising area for further investigation.
The E3 ubiquitin ligase Rnf20 was identified as a potential negative regulator of Foxp3 via CRISPR screen, and therefore a suitable target for immunotherapies [Citation129]. With the advancement of immunotherapy, there is an optimistic outlook for new therapeutic strategies, including those targeting UHRF1 and circUHRF1, which may ultimately improve the prognosis and treatment outcomes for cancer patients. However, continuing investigation and the initiation of clinical trials will be necessary to confirm the practical application of these potential therapies beyond preclinical findings.
Thus, it is reasonable to surmise that numerous RING proteins have the potential to serve as effective therapeutic targets for various diseases. Gaining insight into the functions of RING finger proteins and their associated complexes, including BRCA1–BARD1 [Citation29,Citation30], RNF20–RNF40 [Citation9,Citation44], KDM2B–PCGF1–RING1B [Citation22,Citation23] and CUL4–DDB–RBX1 [Citation16], within chromatin intricacies will help to understand the underlying mechanisms driving cancers and other diseases. It will provide new perspectives that may lead to more precise and effective therapeutic approaches to combat various diseases.
Although a handful of small-molecule inhibitors that target RING finger proteins are currently undergoing clinical trials [Citation122], no drugs targeting these proteins are currently available on the market. Consequently, there is still a substantial need to develop inhibitors that target RING proteins.
Conclusion
This review elucidates the roles and underlying mechanisms of RING finger proteins as E3 ubiquitin ligases in regulating chromatin remodeling by ubiquitinating various histone Lys sites and interacting with other histone PTMs, such as histone methylation and histone acetylation. RING finger protein-mediated histone ubiquitination is involved in regulating gene transcription and DNA damage response concerning numerous specific diseases, such as cancers and autoimmune diseases. Targeting these RING finger proteins and the complexes containing RING finger proteins might be potentially effective therapeutic strategies. Other potential alternatives for targeting RING finger proteins may also help improve therapeutic strategies, such as targeting upstream regulatory proteins of RING finger proteins. Finally, designing inhibitors to target the crosstalk between histone ubiquitination and histone methylation/acetylation may provide research directions for the subsequent development of therapeutic strategies for malignancy.
Future perspective
The potential clinical implications of targeting histone ubiquitination mediated by RING finger proteins are particularly exciting. By selectively modulating specific events of histone ubiquitination, it may be possible to manipulate gene-expression patterns associated with various diseases, including cancers. These processes can potentially open avenues for the development of innovative therapeutic interventions that can restore aberrant gene-expression profiles to a healthier state. Exploring the genetic alterations of RING finger proteins in cancer and other diseases will establish new avenues for future research and provide ideas for discovering new therapeutic strategies. Understanding the intricate crosstalk between histone ubiquitination and other epigenetic modifications, such as methylation and acetylation, has the potential to facilitate combinational therapies that harness synergistic effects. With the advancement of technology allowing for more precise manipulation of histone modifications, there is considerable potential for therapeutic advancements in this area.
Chromatin remodeling
Chromatin remodeling involves switching between a relaxed state (euchromatin) and a more condensed state (heterochromatin).
The loose arrangement of euchromatin facilitates transcription factor binding, leading to increased gene expression. Conversely, the tightly packed heterochromatin limits DNA accessibility, resulting in gene repression.
Post-translational modifications on histones
Histone post-translational modifications (PTMs) mainly include histone methylation, acetylation, ubiquitination and phosphorylation.
PTMs on histones serve as dynamic switches, influencing chromatin compaction and gene accessibility.
RING finger proteins&histone ubiquitination
Histone ubiquitination primarily occurs at core histones and affects gene transcriptional regulation, DNA damage response (DDR) and the maintenance of chromatin integrity.
Histone H2A ubiquitination mainly occurs at lysine (Lys) residues 119/127/129/13/15, contributing to gene repression and DDR.
Histone H2B ubiquitination mainly occurs at Lys-120/34, which promotes transcriptional activation by inhibiting chromatin compaction.
Histone H3 ubiquitination mainly occurs at Lys-14/18/23/36/37/79 and is related to transcriptional regulation, gene stability maintenance, DNA replication and DDR.
Histone H4 ubiquitination mainly occurs at Lys-31/91, facilitating DDR.
RING finger proteins&ATP-dependent chromatin remodeling complexes
RING finger proteins can modulate chromatin structure by ubiquitinating several chromatin-remodeling complexes, including SWI–SNF and FACT.
The intricate interplay between RING finger proteins and chromatin-remodeling complexes has significant implications for regulating gene transcription, DNA damage response and chromatin structure.
Crosstalk between histone ubiquitination&other histone PTMs
The crosstalk between histone ubiquitination and histone methylation is related to gene transcriptional regulation and DDR.
The crosstalk between histone ubiquitination and methylation or acetylation enables the integration of diverse signals and pathways, affording precise control over gene expression in response to alterations in cellular cues.
Biological functions of RING finger proteins in regulating chromatin remodeling
Histone H2AK119 ubiquitination is strongly associated with transcriptional repression, while histone H2B ubiquitination promotes gene transcription.
The ubiquitination of histone H2A cleaves DNA breaks and promotes DNA repair.
Histone H2BK120 monoubiquitination orchestrated by RNF20–RNF40 contributes to the mammalian DDR.
Histone H3 and histone H4 ubiquitination facilitate the recruitment of DDR proteins.
Development of targeted therapeutics for RING finger proteins
BMI1 inhibitor unesbulin/PTC596 has entered phase II clinical trials involving several malignant tumors.
Mithramycin can reduce UHRF1 expression in vitro and in vivo, providing a potential strategy for targeting UHRF1 in malignant pleural mesothelioma.
The E3 ubiquitin ligase Rnf20 may work as a potential target for immunotherapy.
Conclusion
RING finger proteins regulate chromatin remodeling via ubiquitinating different histone lysine sites and interacting with other histone PTMs.
RING finger protein-induced histone ubiquitination participates in gene transcriptional regulation and DDR related to some specific diseases, including cancers and autoimmune diseases.
Designing inhibitors targeting these RING finger proteins or targeting upstream regulatory proteins of RING finger proteins would be an effective therapeutic strategy.
Breaking the crosstalk between histone ubiquitination and other PTMs could be a good option for the development of therapeutic strategies.
Future perspective
Gaining insights into the functions of RING finger proteins and related complexes containing RING finger protein within chromatin intricacies helps to understand the underlying mechanisms driving cancer and other disorders.
Understanding the intricate crosstalk between histone ubiquitination and other epigenetic modifications, such as methylation and acetylation, could pave the way for combinational therapeutic approaches that harness synergistic effects.
Competing interests disclosure
The authors have no competing interests or relevant affiliations with any organization or entity with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Writing disclosure
Medical writing support was provided by LetPub.
Financial disclosure
This study was supported by the HMRF grant (no. 20190152), the Guangdong-Hong Kong Technology Cooperation Funding Scheme (TCFS; no. GHX/092/21SZ) and RGC (no. 14115019). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
- Sullivan BA , KarpenGH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol.11(11), 1076–1083 (2004).
- Kornberg RD , LorchY. Primary role of the nucleosome. Mol. Cell79(3), 371–375 (2020).
- Kouzarides T . Chromatin modifications and their function. Cell128(4), 693–705 (2007).
- Weake VM , WorkmanJL. Histone ubiquitination: triggering gene activity. Mol. Cell29(6), 653–663 (2008).
- Zhang X , LiB , RezaeianAHet al. H3 ubiquitination by NEDD4 regulates H3 acetylation and tumorigenesis. Nat. Commun.8, 14799 (2017).
- Correction to Supporting Information for Chibaya et al. Mdm2 phosphorylation by Akt regulates the p53 response to oxidative stress to promote cell proliferation and tumorigenesis. Proc. Natl Acad. Sci. USA118(9), (2021).
- Bhatnagar S , GazinC , ChamberlainLet al. TRIM37 is a new histone H2A ubiquitin ligase and breast cancer oncoprotein. Nature516(7529), 116–120 (2014).
- Cai C , TangYD , ZhaiJ , ZhengC. The RING finger protein family in health and disease. Signal Transduct. Target. Ther.7(1), 300 (2022).
- Pavri R , ZhuB , LiGet al. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell125(4), 703–717 (2006).
- So CC , RamachandranS , MartinA. E3 ubiquitin ligases RNF20 and RNF40 are required for double-stranded break (DSB) repair: evidence for monoubiquitination of histone H2B lysine 120 as a novel axis of DSB signaling and repair. Mol. Cell Biol.39(8), (2019).
- Nishiyama A , YamaguchiL , SharifJet al. Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature502(7470), 249–253 (2013).
- Jackson SP , DurocherD. Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell49(5), 795–807 (2013).
- Schwertman P , Bekker-JensenS , MailandN. Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat. Rev. Mol. Cell Biol.17(6), 379–394 (2016).
- Hyun K , JeonJ , ParkK , KimJ. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med.49(4), e324 (2017).
- Shvedunova M , AkhtarA. Modulation of cellular processes by histone and non-histone protein acetylation. Nat. Rev. Mol. Cell Biol.23(5), 329–349 (2022).
- Wang H , ZhaiL , XuJet al. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell22(3), 383–394 (2006).
- De Napoles M , MermoudJE , WakaoRet al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell7(5), 663–676 (2004).
- Wang H , WangL , Erdjument-BromageHet al. Role of histone H2A ubiquitination in Polycomb silencing. Nature431(7010), 873–878 (2004).
- Yamamoto Y , AbeA , EmiN. Clarifying the impact of polycomb complex component disruption in human cancers. Mol. Cancer Res.12(4), 479–484 (2014).
- Elderkin S , MaertensGN , EndohMet al. A phosphorylated form of Mel-18 targets the Ring1B histone H2A ubiquitin ligase to chromatin. Mol. Cell28(1), 107–120 (2007).
- Endoh M , EndoTA , ShingaJet al. PCGF6-PRC1 suppresses premature differentiation of mouse embryonic stem cells by regulating germ cell-related genes. Elife6 (2017).
- Wu X , JohansenJV , HelinK. Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol. Cell49(6), 1134–1146 (2013).
- Rona G , RobertiD , YinYet al. PARP1-dependent recruitment of the FBXL10-RNF68-RNF2 ubiquitin ligase to sites of DNA damage controls H2A.Z loading. Elife7 (2018).
- Cao R , TsukadaY , ZhangY. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell20(6), 845–854 (2005).
- Buchwald G , VanDer Stoop P , WeichenriederO , PerrakisA , Van LohuizenM , SixmaTK. Structure and E3-ligase activity of the ring–ring complex of polycomb proteins BMI1 and RING1B. EMBO J.25(11), 2465–2474 (2006).
- Mallette FA , MattiroliF , CuiGet al. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J.31(8), 1865–1878 (2012).
- Pinato S , ScandiuzziC , ArnaudoN , CitterioE , GaudinoG , PenengoL. RNF168, a new RING finger, MIU-containing protein that modifies chromatin by ubiquitination of histones H2A and H2AX. BMC Mol. Biol.10, 55 (2009).
- Kalb R , MalleryDL , LarkinC , HuangJT , HiomK. BRCA1 is a histone-H2A-specific ubiquitin ligase. Cell Rep.8(4), 999–1005 (2014).
- Lorick KL , JensenJP , FangS , OngAM , HatakeyamaS , WeissmanAM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl Acad. Sci. USA96(20), 11364–11369 (1999).
- Wu LC , WangZW , TsanJTet al. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat. Genet.14(4), 430–440 (1996).
- Hashizume R , FukudaM , MaedaIet al. The RING heterodimer BRCA1–BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J. Biol. Chem.276(18), 14537–14540 (2001).
- Starita LM , ParvinJD. Substrates of the BRCA1-dependent ubiquitin ligase. Cancer Biol. Ther.5(2), 137–141 (2006).
- Boulton SJ . BRCA1-mediated ubiquitylation. Cell Cycle5(14), 1481–1486 (2006).
- Barber LJ , BoultonSJ. BRCA1 ubiquitylation of CtIP: just the tIP of the iceberg?DNA Repair (Amst.)5(12), 1499–1504 (2006).
- Chen A , KleimanFE , ManleyJL , OuchiT , PanZQ. Autoubiquitination of the BRCA1*BARD1 RING ubiquitin ligase. J. Biol. Chem.277(24), 22085–22092 (2002).
- Mallery DL , VandenbergCJ , HiomK. Activation of the E3 ligase function of the BRCA1/BARD1 complex by polyubiquitin chains. EMBO J.21(24), 6755–6762 (2002).
- Inoue D , AiharaH , SatoTet al. Dzip3 regulates developmental genes in mouse embryonic stem cells by reorganizing 3D chromatin conformation. Sci. Rep.5, 16567 (2015).
- An JY , KimEA , JiangYet al. UBR2 mediates transcriptional silencing during spermatogenesis via histone ubiquitination. Proc. Natl Acad. Sci. USA107(5), 1912–1917 (2010).
- An JY , KimE , ZakrzewskaAet al. UBR2 of the N-end rule pathway is required for chromosome stability via histone ubiquitylation in spermatocytes and somatic cells. PLOS ONE7(5), e37414 (2012).
- Wang X , KangJY , WeiLet al. Correction: PHF7 is a novel histone H2A E3 ligase prior to histone-to-protamine exchange during spermiogenesis. Development147(8), (2020).
- Zhang Y , MaoD , RoswitWTet al. PARP9-DTX3L ubiquitin ligase targets host histone H2BJ and viral 3C protease to enhance interferon signaling and control viral infection. Nat. Immunol.16(12), 1215–1227 (2015).
- Liu Z , OughtredR , WingSS. Characterization of E3Histone, a novel testis ubiquitin protein ligase which ubiquitinates histones. Mol. Cell Biol.25(7), 2819–2831 (2005).
- Bruhl J , TrautweinJ , SchaferA , LinneU , BouazouneK. The DNA repair protein SHPRH is a nucleosome-stimulated ATPase and a nucleosome-E3 ubiquitin ligase. Epigenetics Chromatin12(1), 52 (2019).
- Zhu B , ZhengY , PhamADet al. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol. Cell20(4), 601–611 (2005).
- Oh S , BooK , KimJet al. The chromatin-binding protein PHF6 functions as an E3 ubiquitin ligase of H2BK120 via H2BK12Ac recognition for activation of trophectodermal genes. Nucleic Acids Res.48(16), 9037–9052 (2020).
- Minsky N , OrenM. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol. Cell16(4), 631–639 (2004).
- Wu L , ZeeBM , WangY , GarciaBA , DouY. The RING finger protein MSL2 in the MOF complex is an E3 ubiquitin ligase for H2B K34 and is involved in crosstalk with H3 K4 and K79 methylation. Mol. Cell43(1), 132–144 (2011).
- Krajewski WA , VassilievOL. Analysis of histone ubiquitylation by MSL1/MSL2 proteins in vitro. Arch. Biochem. Biophys.666, 22–30 (2019).
- Oya E , NakagawaR , YoshimuraYet al. H3K14 ubiquitylation promotes H3K9 methylation for heterochromatin assembly. EMBO Rep.20(10), e48111 (2019).
- Kim CR , NodaT , KimHet al. PHF7 modulates BRDT stability and histone-to-protamine exchange during spermiogenesis. Cell Rep.32(4), 107950 (2020).
- Qin W , WolfP , LiuNet al. DNA methylation requires a DNMT1 ubiquitin interacting motif (UIM) and histone ubiquitination. Cell Res.25(8), 911–929 (2015).
- Li G , JiT , ChenJet al. CRL4(DCAF8) ubiquitin ligase targets histone H3K79 and promotes H3K9 methylation in the liver. Cell Rep.18(6), 1499–1511 (2017).
- Groocock LM , NieM , PruddenJet al. RNF4 interacts with both SUMO and nucleosomes to promote the DNA damage response. EMBO Rep.15(5), 601–608 (2014).
- Fatima A , IrmakD , NoormohammadiAet al. The ubiquitin-conjugating enzyme UBE2K determines neurogenic potential through histone H3 in human embryonic stem cells. Commun. Biol.3(1), 262 (2020).
- Kim K , LeeB , KimJet al. Linker Histone H1.2 cooperates with Cul4A and PAF1 to drive H4K31 ubiquitylation-mediated transactivation. Cell Rep.5(6), 1690–1703 (2013).
- Yan Q , DuttS , XuRet al. BBAP monoubiquitylates histone H4 at lysine 91 and selectively modulates the DNA damage response. Mol. Cell36(1), 110–120 (2009).
- Yang CS , JividenK , SpencerAet al. Ubiquitin modification by the E3 ligase/ADP-ribosyltransferase Dtx3L/Parp9. Mol. Cell66(4), 503–516 e505 (2017).
- Gao Z , LeeP , StaffordJM , Von SchimmelmannM , SchaeferA , ReinbergD. An AUTS2–polycomb complex activates gene expression in the CNS. Nature516(7531), 349–354 (2014).
- Bentley ML , CornJE , DongKC , PhungQ , CheungTK , CochranAG. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. EMBO J.30(16), 3285–3297 (2011).
- McGinty RK , HenriciRC , TanS. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature514(7524), 591–596 (2014).
- Taherbhoy AM , HuangOW , CochranAG. BMI1-RING1B is an autoinhibited RING E3 ubiquitin ligase. Nat. Commun.6, 7621 (2015).
- Gil J , PetersG. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat. Rev. Mol. Cell Biol.7(9), 667–677 (2006).
- Shao Z , RaibleF , MollaaghababaRet al. Stabilization of chromatin structure by PRC1, a polycomb complex. Cell98(1), 37–46 (1999).
- Plath K , FangJ , Mlynarczyk-EvansSKet al. Role of histone H3 lysine 27 methylation in X inactivation. Science300(5616), 131–135 (2003).
- Ross K , SedelloAK , ToddGPet al. Polycomb group ring finger 1 cooperates with Runx1 in regulating differentiation and self-renewal of hematopoietic cells. Blood119(18), 4152–4161 (2012).
- Zhou W , ZhuP , WangJet al. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol. Cell29(1), 69–80 (2008).
- Hu Q , BotuyanMV , ZhaoD , CuiG , MerE , MerG. Mechanisms of BRCA1-BARD1 nucleosome recognition and ubiquitylation. Nature596(7872), 438–443 (2021).
- Mattiroli F , VissersJH , Van DijkWJet al. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell150(6), 1182–1195 (2012).
- Fierz B , ChatterjeeC , McGintyRK , Bar-DaganM , RaleighDP , MuirTW. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat. Chem. Biol.7(2), 113–119 (2011).
- Minsky N , ShemaE , FieldY , SchusterM , SegalE , OrenM. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat. Cell Biol.10(4), 483–488 (2008).
- Machida S , SekineS , NishiyamaY , HorikoshiN , KurumizakaH. Structural and biochemical analyses of monoubiquitinated human histones H2B and H4. Open Biol.6(6), (2016).
- Shema-Yaacoby E , NikolovM , Haj-YahyaMet al. Systematic identification of proteins binding to chromatin-embedded ubiquitylated H2B reveals recruitment of SWI/SNF to regulate transcription. Cell Rep.4(3), 601–608 (2013).
- Densham RM , GarvinAJ , StoneHRet al. Human BRCA1-BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection. Nat. Struct. Mol. Biol.23(7), 647–655 (2016).
- Densham RM , MorrisJR. The BRCA1 ubiquitin ligase function sets a new trend for remodelling in DNA repair. Nucleus8(2), 116–125 (2017).
- Lores P , VisvikisO , LunaR , LemichezE , GaconG. The SWI/SNF protein BAF60b is ubiquitinated through a signalling process involving Rac GTPase and the RING finger protein Unkempt. FEBS J.277(6), 1453–1464 (2010).
- Oliveira DV , KatoA , NakamuraKet al. Histone chaperone FACT regulates homologous recombination by chromatin remodeling through interaction with RNF20. J. Cell Sci.127(Pt 4), 763–772 (2014).
- Liu S , Trejo-ArellanoMS , QiuY , EklundDM , KohlerC , HennigL. H2A ubiquitination is essential for polycomb repressive complex 1-mediated gene regulation in Marchantia polymorpha.Genome Biol.22(1), 253 (2021).
- Blackledge NP , FarcasAM , KondoTet al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell157(6), 1445–1459 (2014).
- Jacquet K , Fradet-TurcotteA , AvvakumovNet al. The TIP60 complex regulates bivalent chromatin recognition by 53BP1 through direct H4K20me binding and H2AK15 acetylation. Mol. Cell62(3), 409–421 (2016).
- Lu LY , WuJ , YeL , GavrilinaGB , SaundersTL , YuX. RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis. Dev. Cell18(3), 371–384 (2010).
- Cao R , WangL , WangHet al. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science298(5595), 1039–1043 (2002).
- Kuzmichev A , NishiokaK , Erdjument-BromageH , TempstP , ReinbergD. Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of Zeste protein. Genes Dev.16(22), 2893–2905 (2002).
- Kim J , GuermahM , McGintyRKet al. RAD6-mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell137(3), 459–471 (2009).
- Hwang WW , VenkatasubrahmanyamS , IanculescuAG , TongA , BooneC , MadhaniHD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell11(1), 261–266 (2003).
- Feng Q , WangH , NgHHet al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol.12(12), 1052–1058 (2002).
- McGinty RK , KimJ , ChatterjeeC , RoederRG , MuirTW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature453(7196), 812–816 (2008).
- Ng HH , XuRM , ZhangY , StruhlK. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem.277(38), 34655–34657 (2002).
- Worden EJ , HoffmannNA , HicksCW , WolbergerC. Mechanism of cross-talk between H2B ubiquitination and H3 methylation by Dot1L. Cell176(6), 1490–1501 e1412 (2019).
- Jang S , KangC , YangHSet al. Structural basis of recognition and destabilization of the histone H2B ubiquitinated nucleosome by the DOT1L histone H3 Lys79 methyltransferase. Genes Dev.33(11–12), 620–625 (2019).
- Yao T , JingW , HuZet al. Structural basis of the crosstalk between histone H2B monoubiquitination and H3 lysine 79 methylation on nucleosome. Cell Res.29(4), 330–333 (2019).
- Anderson CJ , BairdMR , HsuAet al. Structural basis for recognition of ubiquitylated nucleosome by Dot1L methyltransferase. Cell Rep.26(7), 1681–1690 e1685 (2019).
- Hashimoto H , HortonJR , ZhangX , BostickM , JacobsenSE , ChengX. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature455(7214), 826–829 (2008).
- Arita K , AriyoshiM , TochioH , NakamuraY , ShirakawaM. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature455(7214), 818–821 (2008).
- Sharif J , MutoM , TakebayashiSet al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature450(7171), 908–912 (2007).
- Bostick M , KimJK , EstevePO , ClarkA , PradhanS , JacobsenSE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science317(5845), 1760–1764 (2007).
- Karagianni P , AmazitL , QinJ , WongJ. ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol. Cell Biol.28(2), 705–717 (2008).
- Wu W , NishikawaH , FukudaTet al. Interaction of BARD1 and HP1 Is required for BRCA1 retention at sites of DNA damage. Cancer Res.75(7), 1311–1321 (2015).
- Ikura T , TashiroS , KakinoAet al. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol. Cell Biol.27(20), 7028–7040 (2007).
- Zeng S , WangY , ZhangT , BaiL , WangY , DuanC. E3 ligase UHRF2 stabilizes the acetyltransferase TIP60 and regulates H3K9ac and H3K14ac via RING finger domain. Protein Cell8(3), 202–218 (2017).
- Henry KW , WyceA , LoWSet al. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev.17(21), 2648–2663 (2003).
- Kao CF , HillyerC , TsukudaT , HenryK , BergerS , OsleyMA. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev.18(2), 184–195 (2004).
- Lehmann L , FerrariR , VashishtAA , WohlschlegelJA , KurdistaniSK , CareyM. Polycomb repressive complex 1 (PRC1) disassembles RNA polymerase II preinitiation complexes. J. Biol. Chem.287(43), 35784–35794 (2012).
- Black JC , ChoiJE , LombardoSR , CareyM. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol. Cell23(6), 809–818 (2006).
- Zhu P , ZhouW , WangJet al. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol. Cell27(4), 609–621 (2007).
- Jackson SP , BartekJ. The DNA-damage response in human biology and disease. Nature461(7267), 1071–1078 (2009).
- Aymard F , BuglerB , SchmidtCKet al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol.21(4), 366–374 (2014).
- Ismail IH , AndrinC , McDonaldD , HendzelMJ. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J. Cell Biol.191(1), 45–60 (2010).
- Chitale S , RichlyH. Nuclear organization of nucleotide excision repair is mediated by RING1B dependent H2A-ubiquitylation. Oncotarget8(19), 30870–30887 (2017).
- Doil C , MailandN , Bekker-JensenSet al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell136(3), 435–446 (2009).
- Thorslund T , RipplingerA , HoffmannSet al. Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature527(7578), 389–393 (2015).
- Moyal L , LerenthalY , Gana-WeiszMet al. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol. Cell41(5), 529–542 (2011).
- Huyen Y , ZgheibO , DitullioRAJret al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature432(7015), 406–411 (2004).
- Wakeman TP , WangQ , FengJ , WangXF. Bat3 facilitates H3K79 dimethylation by DOT1L and promotes DNA damage-induced 53BP1 foci at G1/G2 cell-cycle phases. EMBO J.31(9), 2169–2181 (2012).
- Kari V , ShchebetA , NeumannH , JohnsenSA. The H2B ubiquitin ligase RNF40 cooperates with SUPT16H to induce dynamic changes in chromatin structure during DNA double-strand break repair. Cell Cycle10(20), 3495–3504 (2011).
- Nakamura K , KatoA , KobayashiJet al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol. Cell41(5), 515–528 (2011).
- Giannattasio M , LazzaroF , PlevaniP , Muzi-FalconiM. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J. Biol. Chem.280(11), 9879–9886 (2005).
- Johnson RE , HendersonST , PetesTD , PrakashS , BankmannM , PrakashL. Saccharomyces cerevisiae RAD5-encoded DNA repair protein contains DNA helicase and zinc-binding sequence motifs and affects the stability of simple repetitive sequences in the genome. Mol. Cell Biol.12(9), 3807–3818 (1992).
- Elserafy M , AbugableAA , AtteyaR , El-KhamisySF. Rad5, HLTF, and SHPRH: a fresh view of an old story. Trends Genet.34(8), 574–577 (2018).
- Ortiz-Bazan MA , Gallo-FernandezM , SaugarI , Jimenez-MartinA , VazquezMV , TerceroJA. Rad5 plays a major role in the cellular response to DNA damage during chromosome replication. Cell Rep.9(2), 460–468 (2014).
- Moldovan GL , D’andreaAD. DNA damage discrimination at stalled replication forks by the Rad5 homologs HLTF and SHPRH. Mol. Cell42(2), 141–143 (2011).
- Miller AK , MaoG , KnicelyBGet al. Rad5 and its human homologs, HLTF and SHPRH, are novel interactors of mismatch repair. Front. Cell Dev. Biol.10, 843121 (2022).
- Motegi A , LiawHJ , LeeKYet al. Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc. Natl Acad. Sci. USA105(34), 12411–12416 (2008).
- Bruhl J , TrautweinJ , SchaferA , LinneU , BouazouneK. The DNA repair protein SHPRH is a nucleosome-stimulated ATPase and a nucleosome-E3 ubiquitin ligase. Epigenetics Chromatin12(1), 52 (2019).
- Krijger PH , LeeKY , WitNet al. HLTF and SHPRH are not essential for PCNA polyubiquitination, survival and somatic hypermutation: existence of an alternative E3 ligase. DNA Repair (Amst.)10(4), 438–444 (2011).
- Rizo A , OlthofS , HanL , VellengaE , DeHaan G , SchuringaJJ. Repression of BMI1 in normal and leukemic human CD34(+) cells impairs self-renewal and induces apoptosis. Blood114(8), 1498–1505 (2009).
- Vaughan RM , KupaiA , RothbartSB. Chromatin regulation through ubiquitin and ubiquitin-like histone modifications. Trends Biochem. Sci.46(4), 258–269 (2021).
- Reardon ES , ShuklaV , XiSet al. UHRF1 Is a novel druggable epigenetic target in malignant pleural mesothelioma. J. Thorac. Oncol.16(1), 89–103 (2021).
- Zhang PF , GaoC , HuangXYet al. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol. Cancer19(1), 110 (2020).
- Cortez JT , MontautiE , ShifrutEet al. CRISPR screen in regulatory T cells reveals modulators of Foxp3. Nature582(7812), 416–420 (2020).
