Welcome to the 16th volume of Epigenomics. I would like to take this opportunity to wish you all a happy new year in 2024. I will be looking back at the journal’s highlights of 2023 in this foreword. In the past year, authors of Epigenomics have continued to advance in biomedical sciences, sharing their latest research and new insights, as well as reviewing important topics on epigenetics. It is our privilege to continue publishing these high-quality articles.
This year Epigenomics saw a change in the editorial team, with myself taking over as the Journal Development Editor from Jack Lodge. I will continue to work with our passionate and committed scientists in the field on their contributions and developments for the journal.
2023 Article highlights
This year we published a themed issue on the topic of “Therapies & non-coding RNA drug discovery in Epigenomics” (Volume 15, Issue 2). This issue shared experts’ opinions on epigenetics in therapeutic strategies for various types of cancers and discussed the potential of translating epigenetics discoveries into clinical benefits, ranging from post-transcriptional ‘gene silencing’ to anti-angiogenic miRNAs.
In 2023, Epigenomics published several different articles with remarkable impacts on this dynamic field, some of which I would like to draw your attention to (). The most-read article published in 2023 was ‘Role of epigenetics in pancreatic ductal adenocarcinoma’, a review by Pandey S et al., from the Department of Surgery, University of Miami [Citation1]. The review highlighted the components in epigenetic machinery that take part in pancreatic ductal adenocarcinoma initiation and development. The article also discussed the feasibility of targeting these components as novel therapeutic strategies.
Table 1. Top 5 articles in the 15th Volume of Epigenomics by downloads, as of October 2023.
Another top article from this year is a research article titled ‘Epigenome-wide association study of serum folate in maternal peripheral blood leukocytes’ written by Fragoso-Bargas N et al. [Citation2]. The group from Oslo University Hospital and the University of Oslo in Norway, together with their collaborators from the USA, revealed an epigenome-wide association study of serum folate in maternal blood. They identified novel CpG sites that were associated with serum folate and other cardiometabolic phenotypes. The next most-read article is also a research article ‘Performance comparisons of methylation and structural variants from low-input whole-genome methylation sequencing’. Our editorial board member Sun Z and co-authors compared the performance of various library preparation and sequencing protocols in cases of low-input DNA [Citation3]. They concluded that EM-sequencing is the most suitable method to detect methylation, SNVs and CNVs from single sequencing.
Our most popular Commentary article this year was written by Raphaële Castagné et al., titled ‘The epigenome as a biological candidate to incorporate the social environment over the life course and generations’. The authors shared their insights that epigenetics and epigenetic inheritance can be potential ways to help tackle social inequalities in health and break generational transmission.
‘Beadchip technology to detect DNA methylation in mouse faithfully recapitulates whole-genome bisulfite sequencing’ is also another top-accessed research article from 2023. Martin EM and her colleagues demonstrated the use of mouse methylation BeadChip technology and its performance is comparable to standard whole-genome bisulfite sequencing [Citation5]. This highlights the potential use of BeadChip technology in future DNA methylation research.
Impact & reach of Epigenomics
Authorship locations
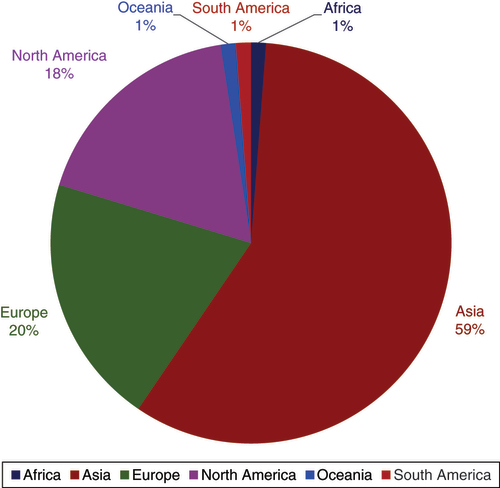
Epigenomics continued to work with authors from all over the world, with Asian, European and North American researchers contributing majorities of the publication in 2023 (). Author’s contribution remained similar compared with 2022. Worthy of note is that authors from Asia rose up by 13% in 2023.
Readership locations
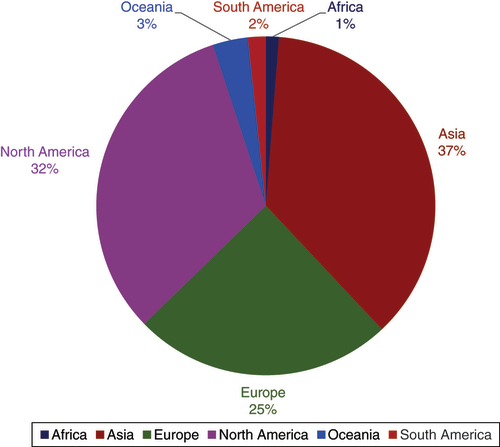
Epigenomics continued to be a globally accessed journal. In 2023, our readers from Asia, North America and Europe remain popularly engaged. The continental ratio for readership is shown below (). We will continue our endeavors to reach audiences from different regions of the world.
Social media
At the time of writing (October 2023), ‘Precision epigenetics provides a scalable pathway for improving coronary heart disease care globally’ by Broyles D and Philibert R [Citation6] gained the highest Altimetric attention score of 54 this year. Their article was reported by seven news outlets and shared broadly among social media. Our journal aims to develop and spread significant and meaningful content available to readers. Epigenomics has a Twitter account (@EpigenomicsFSG) with more than 2000 followers where we share the latest content and updates with the epigenomics community [Citation7]. I invite everyone who wishes to connect with authors and readers to follow us. You are also welcome to join the Epigenomics LinkedIn group and engage with researchers from different parts of the world [Citation8].
Conclusion
On behalf of the editorial team, I would like to express our sincere gratitude to all our authors, peer reviewers and audiences. We also want to extend our gratitude to every member of the Epigenomics Editorial Board for their continuous support. Epigenomics will continue to publish significant research, respected reviews and key opinion articles in 2024. We continue to welcome readers across the globe and consider unsolicited submissions, including original research, editorials, reviews and other article types. For more information regarding submission, please visit our journal website [Citation9].
Future Science Group is committed to making relevant scientific literature accessible to lay audiences. Our Plain Language Summary of Publications are visually enriched articles that provide a summary of a key publication or collection of related clinical studies, and we are delighted to offer this as a publication option in Epigenomics. To find out more about our Plain Language Summary of Publications and how to submit them to the journal, please visit our website [Citation10].
Writing disclosure
No writing assistance was utilized in the production of this manuscript.
Financial disclosure
The author has no financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Competing interests disclosure
M-H Leung is an employee of Future Medicine Ltd, publisher of Epigenomics. The author has no other competing interests or relevant affiliations with any organization or entity with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
- Pandey S , Gupta VK , Lavania SP . Role of epigenetics in pancreatic ductal adenocarcinoma. Epigenomics 15(2), 89–110 (2023).
- Fragoso-Bargas N , Page CM , Joubert BR et al. Epigenome-wide association study of serum folate in maternal peripheral blood leukocytes. Epigenomics 15(1), 39–52 (2023).
- Sun Z , Behati S , Wang P et al. Performance comparisons of methylation and structural variants from low-input whole-genome methylation sequencing. Epigenomics 15(1), 11–19 (2023).
- Castagne R , Ménard S , Delpierre C . The epigenome as a biological candidate to incorporate the social environment over the life course and generations. Epigenomics 15(1), 5–10 (2023).
- Martin EM , Grimm SA , Xu Z , Taylor JA , Wade PA . Beadchip technology to detect DNA methylation in mouse faithfully recapitulates whole-genome bisulfite sequencing. Epigenomics 15(3), 115–129 (2023).
- Broyles D , Philibert R . Precision epigenetics provides a scalable pathway for improving coronary heart disease care globally. Epigenomics 15(16), 805–818 (2023).
- Epigenomics Journal (@EpigenomicsFSG), Twitter. https://twitter.com/EpigenomicsFSG
- Epigenomics, Groups, LinkedIn. www.linkedin.com/groups/8267879/
- Epigenomics . (2023). www.futuremedicine.com/journal/epi
- Future Medicine . Plain Language Summary of Publications. www.futuremedicine.com/plainlanguagesummaries