Abstract
Aim: Mycobacterium avium infections, especially in immune-compromised individuals, present a significant challenge as therapeutic options are limited. In this study, we investigated if M. avium genome encodes nonclassical transpeptidases and if newer carbapenems are effective against this mycobacteria. Materials & methods: Biochemical and microbiological approaches were used to identify and characterize a nonclassical transpeptidase, namely L,D-transpeptidase, in M. avium. Results & conclusion: We describe the biochemical and physiological attributes of a L,D-transpeptidase in M. avium, LdtMav2. Suggestive of a constitutive requirement, levels of LdtMav2, a L,D-transpeptidase in M. avium, remain constant during exponential and stationary phases of growth. Among β-lactam antibacterials, only a subset of carbapenems inhibit LdtMav2 and tebipenem, a new oral carbapenem, inhibits growth of M. avium.
A recent report suggests that global incidence of Mycobacterium avium infections is significant and underestimated [Citation1]. In immune-compromised hosts, for instance, patients infected with HIV, M. avium can cause chronic-disseminated disease with significant morbidity and mortality [Citation2]. M. avium infections are difficult to treat as this pathogen is naturally resistant to most antibacterials available today. Therefore, new insights into vulnerabilities of this pathogen, that may be exploited to develop effective therapies, can have an immediate and a lasting impact.
Mycobacteria possess a large cell wall that accounts for approximately 40% of cell's dry mass [Citation3]. As a major component of the cell wall, the peptidoglycan serves as an anchor to vital cell wall molecules via covalent linkage to arabinogalactan layer to which mycolic acids are associated [Citation4]. Until recently, it was widely accepted that D,D-transpeptidases (commonly known as penicillin-binding proteins) catalyzed the final step of peptidoglycan biosynthesis by linking the fourth amino acid of one stem peptide to the third amino acid of another thereby generating a 4→3 linked 3D-exoskeleton. However, the peptidoglycan of Mycobacterium tuberculosis, Mycobacterium abscessus and Mycobacterium smegmatis are predominantly cross-linked with 3→3 linkages generated by non-classical transpeptidases, namely L,D-transpeptidases [Citation5–9]. Unlike D,D-transpeptidases that use serine to catalyze transpeptidation reaction, the catalytic residue in L,D-transpeptidases is cysteine which when mutated to serine completely abrogates its activity [Citation10]. The predominance of 3→3 cross-linked peptidoglycan in M. tuberculosis is an important reason why β-lactam sub-classes, penicillins and cephalosporins are ineffective against this pathogen [Citation6,Citation11]. Penicillins and cephalosporins are also not considered for treatment of M. avium infection as their minimum inhibitory concentration (MIC90) against this pathogen are approximately 40–160 µg/ml [Citation12] and therefore therapeutically irrelevant. In addition to the native β-lactamase present in M. avium [Citation13], the presence of L,D-transpeptidases could potentially contribute toward natural resistance to penicillins and cephalosporins as this class of β-lactams are not known to be effective against L,D-transpeptidases [Citation10,Citation11]. An M. tuberculosis strain lacking the dominant L,D-transpeptidase, LdtMt2, manifested altered cell and colony morphologies, loss of virulence and enhanced sensitivity to β-lactams [Citation6,Citation14]. Here, we began with genomic approaches to identify LdtMt2 orthologs of M. tuberculosis L,D-transpeptidases in M. avium, and focused on an ortholog of LdtMt2, hereafter referred to as LdtMav2. We used biochemical, biophysical, genetic and microbiological approaches to characterize this protein. We explored localization, abundance and relevance of LdtMav2 to physiologically relevant stresses and characterized its interactions with β-lactams. Although generally not considered for treatment of M. avium infections, data from our study merit further investigation of the therapeutic utility of carbapenems against this pathogen. In particular, our finding that a new carbapenem that is orally bioavailable, namely tebipenem, has potent in vitro activity can have direct impact on the treatment of M. avium infections that are resistant to existing drugs.
Materials & methods
Bacterial strains & growth conditions
M. avium 104, a clinical isolate [Citation15], was used in this study and was grown in Middlebrook 7H9 broth (Becton and Dickinson, Sparks, MD, USA) supplemented with 10% (v/v) oleic acid/albumin/dextrose/catalase (OADC), 0.2% glycerol, 0.05% Tween-80 and 50 µg/ml cycloheximide (7H9 complete medium) at 37°C with constant shaking. To assess response to stress, M. avium was grown in 7H9 complete medium to exponential phase (A600 nm = 0.6), centrifuged, washed once with 7H9 broth and resuspended in 7H9 complete medium alone (control), H2O or 7H9 medium supplemented with the following: 0.05% sodium dodecyl sulfate (SDS) or 2 µg/ml crystal violet or 1 µg/ml tebipenem. M. avium was also incubated in Sauton's medium alone and Sauton's medium supplemented with 40 µM ferric ammonium sulfate. Escherichia coli strains, DH5α and BL21(DE3), were grown in Luria-Bertani (LB) broth or LB agar with appropriate selection drug.
Sequence analysis of LdtMav2
Sequence of LdtMav2 was obtained from the Kyoto Encyclopedia of Genes and Genomes (KEGG) M. avium 104 database [Citation16]. Sequence alignment to compare primary structure of LdtMav2 with its orthologs in other mycobacteria and to infer residues potentially involved in catalysis was undertaken using Multiple Alignment using Fast Fourier Transform (MAFFT) [Citation17]. Furthermore, Clustal X was used to color classify each amino acid. The cladogram construction of the sequences was carried out using MEGA 6.06 [Citation18].
Cloning, overexpression & purification
The N-terminal 24 residues of LdtMav2 were excluded to avoid the likely transmembrane anchor predicted by TMHMM [Citation19] that could hinder purification of the protein. Primers attgccatatggtgggggcagtcgcctgcggcg and caatactcgagtcaggtgttggcgttgcccgc were used to amplify the region-encoding residues 25–403 and cloned into pET28a+TEV to generate an N-terminal H6 tagged LdtMav2. E. coli BL21δ∊3 was used to overexpress LdtMav2, treated with TEV protease to cleave the H6 tag while simultaneously dialyzed to remove imidazole. Gel filtration was carried out using Superdex-200 10/300 GL (GE Healthcare, IL, USA) to obtain monomeric LdtMav2.
Biochemical characterization
The optimum pH for LdtMav2 was determined by assessing absorbance of opening of the β-lactam ring of nitrocefin at 490 nm, molar extinction coefficient of 20,500 M-1cm-1 as described [Citation20]. Reaction mixtures contained 10 µM LdtMav2, 100 µM nitrocefin, 100 mM-modified buffer containing 2-(N-Morpholino)ethanesulfonic acid (MES), 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) and diethanolamine (1:1:1) at pH ranging from 4 to 10 [Citation21] and 0.1 mM TCEP at 25°C in a total volume of 100 µl. To study LdtMav2 reaction kinetics, nitrocefin was used as a substrate at varying concentrations at 25°C. For each reaction, a control in which the rate of nitrocefin hydrolysis in the absence of enzyme at each pH and substrate concentration was included. The Km and Vmax were determined using nonlinear regression analysis of which Michaelis Menton gave the best fit using Graph pad prism (Version 6; La Jolla, CA, USA).
Inactivation by β-lactams & mass spectrometry to identify adduct
To characterize reaction between LdtMav2 and various β-lactam subclasses, enzyme–inhibitor (EI) complex was prepared by incubating LdtMav2 with 200 µM of carbapenem at 25°C for 2 h. Unreacted inhibitor was removed by size exclusion chromatography. The purified acyl-enzyme was incubated with nitrocefin and its hydrolysis was monitored at 495 nM. Nitrocefin incubated with unreacted LdtMav2 was included as a control. To identify adduct formed with LdtMav2, each β-lactam was incubated at 4 mM concentration with 2 µM enzyme in 400 µl of 25 mM Tris, pH 8, for 5 h at room temperature. These samples were analyzed using a Waters Aquity H-Class Ultra Performance Liquid Chromatography–Mass Spectrometry (UPLC–MS) system and adducts were identified as previously described [Citation10].
Determination of kinetic rate constants
The λmax for each β-lactam was determined spectroscopically and they are: amoxicillin, 272 nm; cephalothin, 270 nm; doripenem, 300 nm; faropenem, 306 nm; and T210, 303.5 nm. T210 is an experimental carbapenem synthesized by our group [Citation10]. Extinction coefficients for these compounds were determined for doripenem (7500 M-1 cm-1), biapenem (9845.7 M-1 cm-1), faropenem (6980 M-1 cm-1), tebipenem (6652 M-1 cm-1) and T210 (9880 M-1 cm-1). A stopped-flow apparatus (SX20; Applied Biophysics, NY, USA) was used to measure fluorescence of reaction in sodium phosphate buffer, pH 7.4 at 10°C as described [Citation22]. Fluorescence emission was recorded at 320 nm. Fluorescence data were analyzed using regression analysis using the equation [EI*] = [Etotal](1-e-kobst). [Etotal] is the total enzyme concentration, kobs is the constant to be determined and t is time. The kobs values were plotted versus the concentration of the inhibitor [I] and regression analysis was performed using the equation, kobs = kinact[I]/Kapp + [I], where kinact is a first-order constant for acylenzyme formation and Kapp is a constant.
Abundance & cellular localization
Abundance of LdtMav2 was studied using western blot analysis with polyclonal antibody generated against full-length LdtMav2 in rabbits (Spring Valley Laboratory, MD, USA). M. avium cultures grown to exponential (A600 nm = 0.6) and stationary phases of growth (A600 nm ∼2.4) were used to extract cytosolic and membrane protein fractions, electrophoresed on 10% SDS-PAGE gels and transferred to a polyvinylidene difluoride membrane (GE Healthcare, IL, USA). Following overnight blocking, the membrane was incubated with anti-LdtMav2 polyclonal antibody (1:1000) in Tris-buffered saline (TBS) containing 0.2% bovine serum albumin (BSA) and 0.1% Tween-80 (TBS-T) for 2 h at room temperature and was washed three-times with TBS-T for 15 min and incubated with secondary antirabbit-horseradish peroxidase conjugate (Cell Signaling, MA, USA). The blot was washed three-times with TBS-T, and developed using chemiluminescence. Purified LdtMav2 was included as a control.
Cellular localization of LdtMav2 was studied using cell wall and cytosolic fractions of M. avium prepared from exponential and stationary-phase cultures using a slight modification of a validated protocol [Citation23]. Expression levels of LdtMav2 in M. avium was assessed from whole-cell lysates made using M. avium 104 cultures grown to exponential (A600 nm = 0.6) and stationary phase (A600 nm = 2.4). The cells were centrifuged at 3000 ×g for 10 min at 4°C, the pellet was resuspended in a buffer containing 50 mM Tris-HCl (pH 8), 150 mM NaCl, 0.1 mM TCEP, 1 mM PMSF, 10% glycerol and treated with lysozyme (2 mg/ml overnight at 4°C). This sample was sonicated on an ice–ethanol mixture followed by centrifugation at 3000 ×g for 10 min to remove any unbroken cellular debris. The lysate obtained after sonication was centrifuged at 3000 ×g for 10 min to remove cellular debris and then centrifuged again at 30,000 ×g for 30 min at 4°C. The supernatant from this sample represents the cytosol and cell pellet is the cell wall fraction. The cell lysate proteins after quantification using the modified Bradford method [Citation24] and normalization were electrophoresed on 10% SDS-PAGE gels and transferred to a polyvinylidene difluoride membrane (GE) using a semidry transfer apparatus (Bio-Rad, Hercules, CA, USA). The membrane was blocked using 5% blocking solution nonfat dry milk (Cell Signalling, MA, USA) in the presence of a buffer containing 0.2% BSA in TBS (20 mM Tris-HCl [pH 7.4], with 150 mM NaCl) overnight. Excess blocking agent was removed by washes in a buffer containing TBS (20 mM Tris-HCl [pH 7.4], with 150 mM NaCl) and 0.1% Tween-20. The membrane was then incubated with the polyclonal anti-LdtMav2 antibody (1:1000) in TBS containing 0.2% BSA and 0.1% Tween-80 for 2 h. The membrane was washed three-times with TBS containing 0.05% Tween-20 over 15 min and incubated with secondary antirabbit-horseradish peroxidase conjugate (Cell signaling) at a 1:10,000 dilution for 1 h. The blot was washed three-times with TBS-T, and was developed using chemiluminescence-developing kit (Santa Cruz Biotechnology, TX, USA), following the manufacturer's protocol. Purified MAV_1661 was used as a control.
Protein–protein interaction
A pull-down assay was used to identify any protein(s) that interacted with LdtMav2. M. avium lysate was prepared as described above from cultures harvested at exponential growth phase (A600 nm = 0.6). The lysate was first interacted with Ni-NTA resin at 4°C for 2 h. This precleared lysate was interacted with His-tagged LdtMav2 immobilized on Ni-NTA agarose resin at 4°C for 2 h. The beads were washed with 0.5% Triton-X, 50 mM Tris-HCl (pH 8), 150 mM NaCl, 0.1 mM TCEP, 1 mM PMSF, 10% glycerol and 5 mM imidazole and analyzed using 10% SDS-PAGE gel. The protein band that was exclusively present in the presence of LdtMav2 was excised, digested with trypsin and subjected to mass spectrometric analysis to determine the peptide fragment masses, which were then, analyzed against the MASCOT database to identity the protein.
Minimum inhibitory concentration & checkerboard titration assay
MIC90 of agents against M. avium was determined using the standard broth microdilution assay [Citation25], using 96-well plates separately at pH 6.8 (Middlebrook 7H9 broth in the presence of 10% OADC) and pH 7.4 (cation-adjusted Mueller-Hinton broth) in accord with the Clinical and Laboratory Standard Institute (CLSI) [Citation26] guidelines described in [Citation27]. Standard Checkerboard Titration assay [Citation28] was used to assess combined activity of agents. Two drugs were added to 2.5 ml of Middlebrook 7H9 broth, each starting at its MIC90 and serially diluted twofold, so all possible twofold dilution combinations below respective MICs are represented. Next, 105 CFU of M. avium were inoculated into each tube. The cultures were incubated at 37°C and evaluated for growth by visual inspection of the broth at 9 days as per CLSI guidelines. Fractional inhibitory concentration (FIC) of a drug was calculated as described [Citation28]. FIC of a drug in a sample is computed as concentration of the drug divided by the MIC of the drug when used alone. FIC index is the sum of FIC of two drugs in a sample. FIC index was calculated for each mix of drugs that inhibited M. avium growth and an average FIC index was determined. Average FIC index ≤0.5 was interpreted as synergy, 0.5–2.0 as indifference and more than two as antagonism.
Results
LdtMav2 is conserved across most mycobacteria
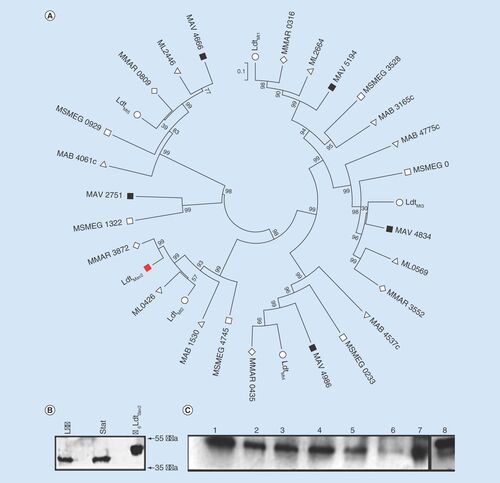
We used LdtMt2, the dominant L,D-transpeptidase in M. tuberculosis [Citation6], to identify its sequence ortholog, LdtMav2, in M. avium 104. According to the existing annotation (accession WP_033706218.1), LdtMav2 is a 379 aa protein encoded by locus MAV_1661 [Citation29]. LdtMav2 shares 86% sequence identity with LdtMt2 and possesses a C-terminal YkuD domain (Pfam 03734) that is characteristic of L,D-transpeptidases and two bacterial immunoglobulin-like domains (Ig) in the N-terminus (Supplementary Figure 1) [Citation30]. While orthologs of LdtMav2 also exist in Mycobacterium leprae (ML0426), M. abscessus (MAB_1530), M. smegmatis (MSMEG_4745), Mycobacterium marinum (MMAR_3872), five additional paralogs are encoded by M. avium 104 genome (). Widespread occurrence of multiple L,D-transpeptidase orthologs across different species of mycobacteria may be indicative of their significance to the cellular physiology of this group of pathogens. As the majority of cross-linkages in the peptidoglycan of M. tuberculosis [Citation5,Citation7] and M. abscessus [Citation8] are generated by L,D-transpeptidases, we hypothesize that the presence of six paralogs in M. avium 104 is suggestive of notable significance of this protein class to the peptidoglycan physiology in this organism.
(A) Cladogram of L,D-transpeptidases from various mycobacteria. Multiple alignment and phylogeny analysis of L,D-transpeptidases from Mycobacterium tuberculosis (O), Mycobacterium smegmatis (□), Mycobacterium absessus (Δ), Mycobacterium leprae (∇), Mycobacterium marinum (◊) and Mycobacterium avium (□). LdtMav2 is highlighted as a red-closed square. (B) Western blot analysis of cell wall and cytosolic fractions of M. avium harvested at logarithmic (log) and stationary (stat) phases of growth and probed with antibody against LdtMav2. His6-tagged LdtMav2 is included as a control (far right). (C) Levels of LdtMav2 in the presence of 100 µM iron (lane 1), 40 µM iron (lane 2), 0.05% SDS (lane 3), 2 µg/ml crystal violet (lane 4), 1 µg/ml tebipenem (lane 5), H2O (lane 6), Middlebrook 7H9-enriched medium (lane 7) and His6-tagged LdtMav2 as a control (lane 8).
LdtMav2 localizes to the cell envelope, interacts with a putative Ser/Thr kinase & responds to stress
We used western blot analysis using an antibody raised against LdtMav2 to probe this protein in M. avium. We observed a single band of similar intensity in samples from both exponential and stationary growth phases but only in the cell wall fraction (). This protein is similar in size to LdtMav2 expressed in and isolated from E. coli that was included as a control. Therefore, we conclude that LdtMav2 is located in the cell wall complex that includes peptidoglycan and its abundance is similar during exponential and stationary phases of growth in vitro in nutrient-rich broth. In addition to the transmembrane anchor domain and the catalytic domain resembling LdtMt2 [Citation30], LdtMav2 also possesses two Ig domains spanning residues 23–119 and 123–221. Functions, if any, of these domains are not known. We hypothesized that Ig domains in LdtMav2 may be involved in interacting with other proteins. To test this hypothesis, we performed a pull-down assay using M. avium whole-cell lysate and analyzed the bound proteins using mass spectrometry. The following four proteins were present in highest abundance: LdtMav2, trypsin, serum albumin and an uncharacterized protein of M. avium (accession # 497665596) with an abundance significance scores of 1593, 866, 305 and 112, respectively. Presence of LdtMav2, trypsin and albumin (that were added during preparation of protein for mass spectrometric analysis) were expected and served as internal controls. Further analysis using BLASTp with the sequence of the bound protein identified the uncharacterized protein that bound to LdtMav2 to be a Ser/Thr kinase (Supplementary Table 1) with a homodimer interface with polypeptide-binding sites.
Given the localization of LdtMav2 in the cell wall and its likely function as a catalyst of the final step of peptidoglycan synthesis, we hypothesized that it may play a role responding to environmental stresses. We probed LdtMav2 levels in the presence of chemical stresses relevant to cell wall physiology or those that mimic stresses present during an infection. For this purpose, we exposed M. avium to low iron environment, osmotic shock, sublethal concentrations of cell wall perturbants such as sodium dodecyl sulfate [Citation31], crystal violet [Citation32] and the peptidoglycan biosynthesis inhibitor tebipenem [Citation33]. While LdtMav2 levels were similar in M. avium grown in the nutrient-rich Middlebrook 7H9 complete medium and Sauton's broth supplemented with 100 µM iron, a small but noticeable decrease was observed in the presence of 40 µM iron, 0.05% sodium dodecyl sulfate and 2 µg/ml crystal violet (). Exposure to 1 µg/ml tebipenem and osmotic shock resulted in very low levels of LdtMav2.
LdtMav2 hydrolyzes β-lactam substrate
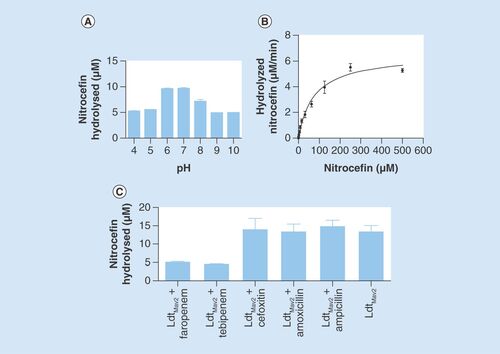
H6-tagged LdtMav2 (25–403 aa fragment) was expressed in the heterologous host E. coli BL21δ∊3, and purified to apparent homogeneity using Ni-NTA chromatography. Further purification and assessment by gel filtration revealed that LdtMav2 is stably present in its monomeric form. Attempts to purify the YkuD domain alone (residues 253–403) were unsuccessful (data not shown), suggesting that perhaps the Ig domains are essential for stabilizing the YkuD fold. L,D-transpeptidases bind to and hydrolyze a tetrapeptide substrate during peptidoglycan biosynthesis [Citation11]. By mimicking the native substrate, β-lactams are also bound and hydrolyzed by L,D-transpeptidases LdtMt1 [Citation34], LdtMt2 [Citation30] and LdtMt5 of M. tuberculosis [Citation20]. Given extensive sequence similarity to LdtMt2, we hypothesized that LdtMav2 would also bind to and catalyze hydrolysis of β-lactams. We used nitrocefin, a chromogenic β-lactam, as a substrate and observed that hydrolytic activity of LdtMav2 peaks in the physiological pH range (). LdtMav2 displayed a classical Michaelis–Menten kinetics for hydrolysis of nitrocefin with Km and Vmax of 75 µM and 6.5 μM/min, respectively. Additionally the kinetic parameters with respect to hydrolysis of nitrocefin are comparable to LdtMt2 [Citation30] and LdtMt5 [Citation20]. The rate of nitrocefin hydrolysis was a slow reaction with a Kcat of 0.65 ± 0.03 min-1. The efficiency of LdtMav2 catalysis of nitrocefin was calculated as a Kcat/Km ratio of 8.7 × 103 min-1M-1 ().
(A) pH profile of nitrocefin hydrolysis by LdtMav2. (B) Rate of nitrocefin hydrolysis by LdtMav2 at pH 6.5. (C) Assessment of nitrocefin hydrolysis by LdtMav2 following incubation with faropenem, tebipenem, cefoxitin, amoxicillin and ampicillin. The experiments were performed three-times and error bars represent standard error.
Carbapenems & cephalothin irreversibly inhibit LdtMav2
Although β-lactams bind to and inhibit D,D-transpeptidases [Citation35], it is not known if this antibacterial class interacts with LdtMav2. We studied the nature of the reaction between LdtMav2 and different β-lactam subclasses using UPLC-MS. Amoxicillin and ampicillin were used as representatives of penicillin subclass; cephalothin and cefoxitin are cephalospirins; and doripenem, biapenem and tebipenem are carbapenems while faropenem is a penem. While cephalothin, imipenem, doripenem, biapenem, faropenem and tebipenem formed covalent acyl-adducts, we were unable to detect any reacted masses for amoxicillin, ampicillin, cefoxitin and meropenem (). Additionally, we used nitrocefin as a substrate to monitor LdtMav2 activity following treatment with the β-lactams. Hydrolysis of nitrocefin by LdtMav2 was significantly inhibited when the enzyme was treated with faropenem or tebipenem (). Cefoxitin, amoxicillin and ampicillin failed to inhibit the enzyme as nitrocefin hydrolysis occurred despite treatment with these agents.
Kinetics of acylation of LdtMav2
Kinetics of LdtMav2 acylation by various β-lactams was assessed in the presence of increasing concentrations of β-lactams and a fixed concentration of LdtMav2. Parameters that characterize the kinetics of reaction, kinact and Kapp were determined were determined to be as follows: doripenem (0.7 s-1 and 14 μM), biapenem (0.6 s-1 and 17 μM), faropenem (0.4 s-1 and 8 μM), tebipenem (0.3 s-1 and 13 μM), cephalothin (0.4 s-1 and 35 μM) and T210 (0.4 s-1 and 1.7 μM; ). The differences in kinetics reflect changes in the chemistry of binding among the β-lactams. Amoxicillin (Supplementary Figure 2) and cefoxitin (data not shown) failed to inhibit LdtMav2. The kinact/Kapp ratios for doripenem (0.5 s-1 μM-1), biapenem (0.04 s-1 μM-1), faropenem (0.5 s-1 μM-1), tebipenem (0.02 s-1 μM-1), cephalothin (0.1 s-1 μM-1) and T210 (0.2 s-1 μM-1) illustrate that the efficiencies of LdtMav2 inhibition vary significantly among β-lactams.
Plots of kobs as a function of concentration of various carbapenems. Regression analysis was performed using the equation, kobs = kinact[I]/Kapp + [I], where kinact is a first-order constant for acylenzyme formation, and Kapp is a constant. The experiments were performed at least thrice and error represents standard deviation.
![Figure 3. Kinetics of β-lactam ring opening by LdtMav2.Plots of kobs as a function of concentration of various carbapenems. Regression analysis was performed using the equation, kobs = kinact[I]/Kapp + [I], where kinact is a first-order constant for acylenzyme formation, and Kapp is a constant. The experiments were performed at least thrice and error represents standard deviation.](/cms/asset/fecc1532-4d7d-487f-a36a-46f8c0d2bcb6/ifmb_a_12327487_f0003.jpg)
M. avium is susceptible to tebipenem but resistant to most carbapenems
We assessed a panel of β-lactams, including newer carbapenems, for activity against M. avium according to CLSI guidelines [Citation26]. Tebipenem, a recently developed carbapenem with oral bioavailability and potency against M. tuberculosis [Citation33], exhibited MIC90 of 2–4 µg/ml at pH 6.8 and 4–8 µg/ml in the pH 7.4 and was the only β-lactam (among those tested in this study) with a potency in the therapeutically relevant range. Other carbapenems failed to inhibit M. avium growth even at 64 µg/ml, the highest concentration in the range tested (). Next, we supplemented each of these β-lactams with clavulanate, 5 µg/ml, to assess if this β-lactamase inhibitor enhanced the susceptibility of M. avium to the β-lactams: the MIC of the β-lactams were not altered by the addition of clavulanate.
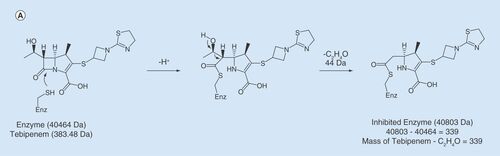
Incubation of LdtMav2 with tebipenem (Molecular weight; MW = 383.5 Da) produced a covalently bound acyl adduct of +339 Da (). The catalytic domain of LdtMt2 and the homologous domain in LdtMav2 share 96% sequence identity (Supplementary Figure 1). In LdtMt2, catalytic site residues Tyr318, His332, His352 and Cys354 are involved in coordination and reaction with carbapenems [Citation36]. These residues are conserved in LdtMav2. Based on these data, we propose the following mechanism for tebipenem adduct formation with LdtMav2. Sulfur atom of the putative catalytic Cys349 makes a nucleophilic attack to the carbonyl carbon of β-lactam ring and opens the ring (). Next, as in meropenem and tebipenem reactions with LdtMt2 [Citation36], hydroxyethyl group at C6 is lost resulting in an adduct of +339 Da.
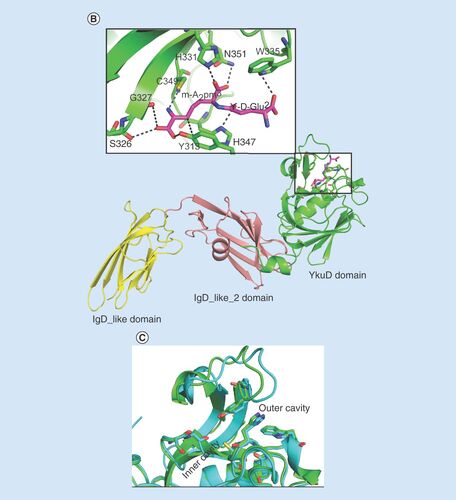
(A) Proposed mechanism of acylation of LdtMav2 by tebipenem. (B) Model of LdtMav2 with different domains, Ig-like domain (yellow), Ig-like_2 domain (salmon) and YkuD domain (green). The inset picture shows the active site of LdtMav2 with peptidoglycan substrate fragment γ-D-Glu-m-A2pm dipeptide (pink) showing interactions with different residues. (C) Superposition of active site of LdtMav2 (green) with LdtMt2 (cyan).
Carbapenems and rifampin exhibit synergy in vitro against M. tuberculosis and M. abscessus [Citation27]. We hypothesized that tebipenem may exhibit synergy with rifampin against M. avium. Checker board assay revealed a lack of synergy between tebipenem and rifampin against this mycobacterium (Supplementary Table 1). Clarithromycin and moxifloxacin are commonly used for treatment of M. avium infections [Citation37]. We also studied potencies of combination of clarithromycin or moxifloxacin and tebipenem; these combinations failed to exhibit synergy against M. avium (Supplementary Table 1).
Molecular modeling of LdtMav2 with putative substrate
Structural characterization of LdtMav2 can provide further insight into its biochemical properties. Using the crystal structure of LdtMt2 with a native peptide substrate bound to its catalytic site (PDB ID: 3TUR) [Citation30] as a template, we used homology modeling to generate a 3D model of LdtMav2 (). The modeling study suggests that overall structure of LdtMav2 is very similar to LdtMt2. The three domains, namely IgD1, IgD2 and the YkuD domain, share very similar structures and topologies. The peptidoglycan substrate fragment γ-D-Glu-m-A2pm dipeptide modeled through the outer cavity into the active site of LdtMav2 shows interactions with putative active-site residues including His331, Trp335, Tyr313, His347, Gly327, Asn351 and Cys349. Superposition of active sites of LdtMav2 with LdtMt2 () reveals that residues involved in substrate binding are conserved in the two proteins, suggesting a similar mechanism of catalysis and substrate specificities.
Discussion
Mycobacterial genomes encode multiple sequence orthologs with the YkuD domain (). The presence of this domain is predictive of a function related to L,D-transpeptidation. While a recent study ascribed L,D-transpeptidase function to the paralogs in M. tuberculosis using an in vitro peptide cross-linking assay [Citation38], studies that evaluated strains lacking each paralog concluded them to be functionally nonredundant [Citation6,Citation14,Citation20]. The presence of six sequence orthologs in M. avium genome illustrates that a significant catalytic potential of this protein class is preserved by this pathogen and perhaps also suggests their importance to this organism. In this study, we have chosen to characterize LdtMav2, whose ortholog in M. tuberculosis (LdtMt2) is the dominant L,D-transpeptidase [Citation6].
As β-lactams are known to mimic peptide substrates of peptidoglycan biosynthesis [Citation35] and LdtMav2 readily hydrolyzed nitrocefin, a β-lactam, it is reasonable to expect that this enzyme uses peptides or related biomolecule as its native substrate. Localization of LdtMav2 in the cell wall further suggests that similar to its ortholog LdtMt2, it is likely anchored to the cell membrane with a single transmembrane domain and catalyzes formation of 3→3 linkages in the peptidoglycan layer. We found that LdtMav2 associates with a putative Ser/Thr kinase (accession #: 48928156) possessing a homodimer interface for peptide-binding site. These two proteins exist at the site of peptidoglycan synthesis. Therefore, it is possible that the homodimer interface interacts with the stem peptide fragments generated and released during peptidoglycan metabolism and the Ser/Thr kinase domain of the protein transmits this signal to LdtMav2 or another protein. There is a precedent to this mechanism: in Bacillus subtilis a conserved membrane bound Ser/Thr kinase binds to peptidoglycan fragments and regulates spore germination [Citation39]. The presence of similar levels of LdtMav2 during exponential and stationary phases of growth suggests that activity conferred by this protein is likely required constitutively by M. avium. Lower abundance of the protein, especially when M. avium is exposed to osmotic shock or tebipenem, suggests that this organism may also deploy alternative mechanisms to fortify its cell wall.
Treatment of infections with drug-resistant strains of M. avium pose serious challenges as the limited choices of antibacterials are often not well tolerated by patients. Therefore, availability of new and well-tolerated treatment agent that functions by inhibiting previously unexploited target can have a direct impact on the health of patients with M. avium infection. Consistent with the recent observations that M. tuberculosis L,D-transpeptidases are irreversibly inhibited by carbapenems [Citation36,Citation38], we also observed that stable covalent adducts are formed between LdtMav2 and carbapenems. The efficiency of acylation of LdtMav2, as can be described by kinact/Kapp, shows similar acylation kinetics among the carbapenems. However, this in vitro inhibition did not completely predict whole-cell activity: although all tested carbapenems except meropenem covalently acylated LdtMav2, only tebipenem exhibited whole-cell potency against M. avium. Several possibilities can be speculated to account for the apparent difference between in vitro and whole-cell activities. We hypothesize that tebipenem may be uniquely resistant to hydrolytic activity of the native β-lactamase in M. avium. Equally likely is that tebipenem selectively inhibits other transpeptidases in addition to LdtMav2. The most interesting observation with immediate clinical relevance is that tebipenem not only inhibits LdtMav2 but is also highly potent against M. avium. Therefore, this orally bioavailable antibacterial, from a class namely carbapenems that are generally well tolerated, has the potential to be repurposed as a much needed new therapy to treat M. avium infections. Studies herein, however, do not directly prove if inhibition of LdtMav2 by tebipenem is sufficient to dictate susceptibility of M. avium to this carbapenem.
Conclusion
M. avium genome also encodes for nonclassical transpeptidases, namely L,D-transpeptidases that generate unique 3→3 cross-linkages in the peptidoglycan layer. Tebipenem, a new orally bioavailable carbapenem, not only inhibits M. avium L,D-transpeptidase but also potently inhibits the growth of this pathogen, thereby raises the possibility of using this potent antibiotic to treat conditions arising from infection with this organism.
Future perspective
M. avium infections are rapidly emerging as significant threats in immune-compromised patients as there are very limited antimicrobials that can be used to treat this disease. There is a critical knowledge gap in vital molecular pathways in M. avium that can be targeted to inhibit its growth and leverage this to identify effective antimicrobials. Here, by showing that this pathogen possesses a panel of unexploited enzymes involved in peptidoglycan synthesis and by showing that a commercially available oral drug, namely tebipenem, not only inhibits the enzyme but also inhibits growth of this bacteria, we open the possibility to repurposing this potent antibacterial for treatment of M. avium infections in the future. This study focused on biochemical characterization of a putative L,D-transpeptidase LdtMav2. Availability of molecular tools for efficient manipulation of M. avium would enable genetic studies of L,D-transpeptidases to furnish insight into phenotypes associated with this class of enzymes.
Table 1. Adducts formed between LdtMav2 and β-lactam antibacterials.
Table 2. Potency of various drugs against Mycobacterium avium.
Mycobacterium avium genome encodes six proteins that are sequence paralogs of L,D-transpeptidases that are known to catalyze the nonclassical linkages, namely 3→3 linkages, in the peptidoglycan layer of mycobacteria.
One of the enzymes studied here, LdtMav2, hydrolyzes β-lactam substrates and is also inhibited by certain classes of β-lactams, namely cephalosporins and carbapenems.
One of the carbapenems, namely tebipenem, not only inhibits the enzyme but also has potent activity against M. avium raising the hope that this new orally bioavailable carbapenem may be useful to treat M. avium infections.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Additional file 2
Download MS Word (3 MB)Additional file 3
Download MS Word (165.9 KB)Supplementary Figure 1
Download MS Word (14.1 KB)Financial & competing interests disclosure
The authors of this manuscript were supported by the Willowcroft Foundation and the NIH (grant number DP2OD008459). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Additional information
Funding
References
- Raju RM , RajuSM, ZhaoY, RubinEJ. Leveraging advances in tuberculosis diagnosis and treatment to address nontuberculous mycobacterial disease.Emerg. Infect. Dis.22 (3), 365–369 (2016).
- Benator DA , GordinFM. Nontuberculous mycobacteria in patients with human immunodeficiency virus infection.Sem. Respir. Infect.11 (4), 285–300 (1996).
- Goren MB , BrennanPJ. Mycobacterial lipids: chemistry and biologic activities. In : Tuberculosis. YoumansGP( Ed. ). WB Saunders, PA, USA, 63–193 (1979).
- Mahapatra S , BasuJ, BrennanPJ, CrickDC. Structure, biosynthesis, and genetics of the mycolic acid-arabinogalactan-peptidoglycan complex. In : Tuberculosis and the Tubercle Bacillus. ColeST, EisenachKD, McmurrayDN, JacobsWRJr( Eds ). American Society for Microbiology, DC, USA, 275–285 (2005).
- Lavollay M , ArthurM, FourgeaudMet al. The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by L,D-transpeptidation. J. Bact. 190 (12), 4360–4366 (2008).
- Gupta R , LavollayM, MainardiJL, ArthurM, BishaiWR, LamichhaneG. The Mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin.Nat. Med.16 (4), 466–469 (2010).
- Kumar P , AroraK, LloydJRet al. Meropenem inhibits D,D-carboxypeptidase activity in Mycobacterium tuberculosis. Mol. Micro. 86 (2), 367–381 (2012).
- Lavollay M , FourgeaudM, HerrmannJLet al. The peptidoglycan of Mycobacterium abscessus is predominantly cross-linked by L,D-transpeptidases. J. Bact. 193 (3), 778–782 (2011).
- Wietzerbin J , DasBC, PetitJF, LedererE, Leyh-BouilleM, GhuysenJM. Occurrence of D-alanyl-(D)-meso-diaminopimelic acid and meso-diaminopimelyl-meso-diaminopimelic acid interpeptide linkages in the peptidoglycan of Mycobacteria.Biochemistry13 (17), 3471–3476 (1974).
- Kumar P , KaushikA, LloydEPet al. Non-classical transpeptidases yield insight into new antibacterials. Nat. Chem. Biol. 13 (1), 54–61 (2017).
- Mainardi JL , FourgeaudM, HugonnetJEet al. A novel peptidoglycan cross-linking enzyme for a β-lactam-resistant transpeptidation pathway. J. Biol. Chem. 280 (46), 38146–38152 (2005).
- Mizuguchi Y , OgawaM, UdouT. Morphological changes induced by beta-lactam antibiotics in Mycobacterium avium-intracellulare complex.Antimicrob. Agents Chemother.27 (4), 541–547 (1985).
- Prabhakaran K , HarrisEB, RandhawaB, HastingsRC. β-Lactamase activity in mycobacteria including Mycobacterium avium and suppression of their growth by a β-lactamase-stable antibiotic.Microbios81 (328), 177–185 (1995).
- Schoonmaker MK , BishaiWR, LamichhaneG. Nonclassical transpeptidases of Mycobacterium tuberculosis alter cell size, morphology, the cytosolic matrix, protein localization, virulence, and resistance to β-lactams.J. Bact.196 (7), 1394–1402 (2014).
- Horan KL , FreemanR, WeigelKet al. Isolation of the genome sequence strain Mycobacterium avium 104 from multiple patients over a 17-year period. J. Clin. Microb. 44 (3), 783–789 (2006).
- Kanehisa M , SatoY, KawashimaM, FurumichiM, TanabeM. KEGG as a reference resource for gene and protein annotation.NAR44 (D1), D457–D462 (2016).
- Katoh K , MisawaK, KumaK, MiyataT. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform.NAR30 (14), 3059–3066 (2002).
- Tamura K , StecherG, PetersonD, FilipskiA, KumarS. MEGA6: molecular evolutionary genetics analysis version 6.0.Mol. Biol. Evol.30 (12), 2725–2729 (2013).
- Krogh A , LarssonB, Von HeijneG, SonnhammerEL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes.J. Mol. Biol.305 (3), 567–580 (2001).
- Brammer Basta LA , GhoshA, PanYet al. Loss of a functionally and structurally distinct L,D-Transpeptidase, LdtMt5, compromises cell wall integrity in Mycobacterium tuberculosis. J. Biol. Chem. 290 (42), 25670–25685 (2015).
- Newman J . Novel buffer systems for macromolecular crystallization.Acta Crystallogr. D Biol. Crystallogr.60 (Pt 3), 610–612 (2004).
- Triboulet S , ArthurM, MainardiJLet al. Inactivation kinetics of a new target of β-lactam antibiotics. J. Biol. Chem. 286 (26), 22777–22784 (2011).
- Song H , SandieR, WangY, Andrade-NavarroMA, NiederweisM. Identification of outer membrane proteins of Mycobacterium tuberculosis.Tuberculosis (Edinb.)88 (6), 526–544 (2008).
- Zor T , SelingerZ. Linearization of the Bradford protein assay increases its sensitivity: theoretical and experimental studies.Anal. Biochem.236 (2), 302–308 (1996).
- Gavan TL , TownMA. A microdilution method for antibiotic susceptibility testing: an evaluation.Am. J. Clin. Path.53 (6), 880–885 (1970).
- Desmond E . Susceptibility testing of Mycobacteria, Nocardiae and other aerobic Actinomycetes. Clinical Laboratory Standard Institute M24–A2 (2011). http://shop.clsi.org/.
- Kaushik A , MakkarN, PandeyP, ParrishN, SinghU, LamichhaneG. Carbapenems and rifampin exhibit synergy against Mycobacterium tuberculosis and Mycobacterium abscessus.Antimicrob. Agents Chemother.59 (10), 6561–6567 (2015).
- Hsieh MH , YuCM, YuVL, ChowJW. Synergy assessed by checkerboard. A critical analysis.Diagn. Microbiol. Infect. Dis.16 (4), 343–349 (1993).
- Machowski EE , SenzaniS, EalandC, KanaBD. Comparative genomics for mycobacterial peptidoglycan remodelling enzymes reveals extensive genetic multiplicity.BMC Microbiol.14, 75 (2014).
- Erdemli SB , GuptaR, BishaiWR, LamichhaneG, AmzelLM, BianchetMA. Targeting the cell wall of Mycobacterium tuberculosis: structure and mechanism of L,D-transpeptidase 2.Structure20 (12), 2103–2115 (2012).
- Woldringh CL , Van ItersonW. Effects of treatment with sodium dodecyl sulfate on the ultrastructure of Escherichia coli.J. Bact.111 (3), 801–813 (1972).
- Hoffmann CE , RahnO. The bactericidal and bacteriostatic action of crystal violet.J. Bact.47 (2), 177–186 (1944).
- Horita Y , MaedaS, KazumiY, DoiN. In vitro susceptibility of Mycobacterium tuberculosis isolates to an oral carbapenem alone or in combination with β-lactamase inhibitors.Antimicrob. Agents Chemother.58 (11), 7010–7014 (2014).
- Dubee V , TribouletS, MainardiJLet al. Inactivation of Mycobacterium tuberculosis L,D-transpeptidase LdtMt(1) by carbapenems and cephalosporins. Antimicrob. Agents Chemother. 56 (8), 4189–4195 (2012).
- Walsh C . Antibiotics: Actions, Origins, Resistance. ASM Press, DC, USA, 3 (2003).
- Kim HS , KimJ, ImHNet al. Structural basis for the inhibition of Mycobacterium tuberculosis L,D-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains. Acta Crystallogr. D Biol. Crystallogr. 69 (Pt 3), 420–431 (2013).
- Brown-Elliott BA , NashKA, WallaceRJJr. Antimicrobial susceptibility testing, drug resistance mechanisms, and therapy of infections with nontuberculous mycobacteria.Clin. Microbiol. Rev.25 (3), 545–582 (2012).
- Cordillot M , DubeeV, TribouletSet al. In vitro cross-linking of peptidoglycan by Mycobacterium tuberculosis L,D-transpeptidases and inactivation of these enzymes by carbapenems. Antimicrob. Agents Chemother. 57 (12), 5940–5945 (2013).
- Shah IM , LaaberkiMH, PophamDL, DworkinJ. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments.Cell135 (3), 486–496 (2008).
