Abstract
Aims: As part of Singapore’s One Health antimicrobial resistance (AMR) management, this work was designed to understand the AMR burden in recreational beach waters using extended-spectrum beta-lactamase Escherichia coli (ESBL-EC) as an indicator. Materials & methods: A total of 90 water samples were collected from six different recreational beaches over three different time periods. Only 28/90 (31.3%) water samples yielded E. coli colonies ranging from 1 to 80 colony-forming units/100 ml. Results & conclusion: Screening of all colonies using CHROMID® ESBL agar and Luria-Bertani broth supplemented with ceftriaxone showed that none was ESBL-EC. Further monitoring is required to understand the prevalence of ESBL-EC spatiotemporally, contributing to the national AMR surveillance program and providing timely risk assessment for exposure to ESBL-EC.
Extended-spectrum β-lactamases (ESBL) are serine β-lactamases belonging to the class A Ambler molecular and structural classification. These enzymes are characterized by their ability to hydrolyze expanded spectrum β-lactam antibiotics, conferring antimicrobial resistance (AMR) to bacterial strains producing them. ESBL-producing Escherichia coli (ESBL-EC) is a global concern as it causes significant morbidity and mortality in humans [Citation1], exhibits resistance to multiple treatment drugs and is distributed worldwide among humans, animals, food and the environment [Citation2]. It has thus been identified as an indicator organism by the WHO for integrated, multisectoral surveillance of AMR [Citation3].
The burden of ESBL-EC varies across countries, presumably dependent on the availability and effectiveness of antimicrobial policies such as AMR surveillance and restricting access to and judicious use of antimicrobials [Citation4]. In Europe, approximately 15% of E. coli in nosocomial settings were ESBL-EC while in North America, the rate was 13.3% in 2016 [Citation5]. In contrast, the detection rate was over 30% in the Middle East/Africa and in Latin America [Citation5] and more than 40% in Southeast Asia [Citation6]. Similar trends in detection rates were also reported in the community [Citation7].
In Singapore, Tan et al. found that 19.6% of clinical E. coli isolates screened in participating public hospitals during 2006–2007 were ESBL-EC [Citation8]. An increase in the rate of ESBL-EC carriage in the community has also been observed. In 2006, 6.3% of patients that attended the emergency department and had no previous healthcare contact were ESBL-EC carriers [Citation9]. Mo et al. further showed that ESBL-producing Enterobacterales were detected in 26.2% of the gut microbiota of healthy Singaporeans, 88.8% of which were E. coli [Citation10]. Screening of fecal samples from healthy individuals from the Singapore Integrative Omics Study collected in 2018 showed a similar proportion of ESBL-producing Enterobacterales carriage (25.7%), where E. coli was also found to be the most prevalent species [Citation11].
It was hypothesized that the carriage of ESBL-producing Enterobacterales including ESBL-EC may be due to the consumption of contaminated food, travel or exposure to livestock or the environment [Citation12,Citation13]. Studies in Norway and The Netherlands have suggested the risk of ESBL-EC colonization through exposure to recreational beach water or urban aquatic environments [Citation12,Citation14] that may have been contaminated with fecal AMR bacteria through runoff of manure from farms, droppings of animals or the discharge of overflowed treated or untreated sewage during heavy rainfall [Citation15]. However, in countries with closed sewage treatment systems such as Singapore, other sources (e.g., beachgoers or animal waste) may result in contamination of these waters. ESBL-EC isolates from surface waters were found to be more closely related to those isolated from the general populations than isolates from livestock or food reservoirs, further suggesting the likelihood of epidemiological linkage [Citation16].
In Singapore, ESBL-EC have been reported in the Jurong Lake water reservoir and aquaculture sites. Nine ESBL-EC isolated from two random sites at the water reservoir were found to carry the ESBL-encoding genes blaCTX-M-8, blaCTX-M-14, blaCTX-M-15, blaCTX-M-27 and blaCTX-M-55 [Citation17]. ESBL-EC were also detected in surface water and sediments collected from fish farms and along transects that were 100 m away from the farm sites. The prevalence of ESBL-EC was higher in fish farms than in the transect sites (27% vs 1% of the isolated E. coli) [Citation18]. The detection of ESBL-EC in water bodies highlights the potential risk of ESBL-EC exposure to humans. Hence, it is important to survey the prevalence of ESBL-EC in recreational beach waters, where human contact with water bodies is most likely to occur. This study is the first to explore and understand the prevalence of E. coli and ESBL-EC in water samples collected from six of seven popular recreational beaches in Singapore across three days [Citation19].
Materials & methods
Sample collection
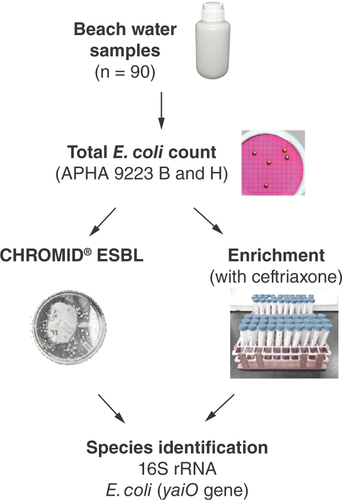
Water from six beaches was sampled, henceforth referred to as Beaches A–F. Beaches A and B are located in the South, Beaches C and D in the East and Beaches E and F in the North of Singapore, respectively (). The number of samples collected from each beach was determined based on the length of the beach where water sample was collected every 500–1000 m along the length of the beach to ensure good coverage. These added up to a total of 30 water samples collected for each sampling session comprising Beach A (n = 12), Beach B (n = 6), Beach C (n = 4), Beach D (n = 4), Beach E (n = 2) and Beach F (n = 2). Each beach was sampled on 3 days, about 3 days to 1 week apart, over a 1-month period in March 2021. Beach water samples were collected by grab sampling during low tide and at a location where the depth of the water is at least 1 m. Before each collection, a waiting time of 1 min was observed to allow any sedimentation to settle. Sample bottles were submerged to 20 cm below the water surface, opened and filled with beach water. Once full, the sample bottle was capped underwater and brought back to shore. The sample bottle was then wiped down with 80% (v/v) ethanol before being stored at 4 °C for transport to the laboratory. The collected samples were then used to enumerate E. coli according to the APHA 9223 B & H standard membrane-filtration method.
Table 1. Sampling coordinates of six beaches and total Escherichia coli count.
Selection & confirmation of ESBL-EC using CHROMID® ESBL agar
Presumptive E. coli isolates were selected based on colony morphology and subcultured onto MacConkey agar and CHROMID® ESBL (bioMérieux, Marcy-l’Etoile, France). A known ESBL E. coli isolate and Klebsiella pneumoniae ATCC 700603 were used to ensure the agar plates produced the expected growth and colony appearance. Following 20–24 h incubation at 37 °C, pink/burgundy colonies noted on CHROMID® ESBL suggest ESBL-EC. These colonies were selected for confirmation using PCR to determine the presence of the yaiO gene as described by Molina et al. () [Citation20]. Briefly, a loop of each colony was resuspended in 100 μl of nuclease-free water and mixed thoroughly. The suspension was boiled at 98 °C for 10 min before centrifugation in a microfuge at maximum speed for 10 min; 1 μl of the supernatant was used as a DNA template for PCR amplification using the Phusion Flash High-Fidelity (HF) PCR Master Mix (Thermo Fisher Scientific, MA, USA). Next, 1 μl of each of the forward and reverse primers (10 μM) was added to the PCR reaction mix and water was added to a final volume of 20 μl. The following PCR amplification conditions were performed: initial denaturation at 98 °C for 10 s; 35 cycles of denaturing at 98 °C for 1 s, annealing at 58 °C for 5 s, extension at 72 °C for 5 s; and final extension at 72 °C for 1 min before an indefinite hold at 16 °C. The PCR products were visualized on a GelRed® (Biotium, CA, USA)-stained 2% TBE agarose gel. Extracted E. coli ATCC 25922 DNA was also used as a positive control for PCR assays.
Species identification was carried out for all colonies grown on CHROMID® ESBL agar by sequencing the 16s rRNA gene using universal primer pairs 27F/1492R () [Citation21]. Briefly, PCR was performed using Phusion Flash HF DNA polymerase in a 20 μl reaction with the addition of 0.2 μl of 10 mM MgCl2, 0.4 μl of each primer (10 μM) and 1 μl of the DNA template that was extracted previously. The PCR assay was performed using the following conditions: initial denaturation at 98 °C for 30 s; 35 cycles of denaturing at 98 °C for 10 s, annealing at 58 °C for 30 s, extension at 72 °C for 60 s; and a final extension at 72 °C for 10 min before an infinite hold at 16 °C. The PCR products were visualized on a GelRed-stained 1% TBE agarose gel. Next, PCR products were purified using Expin PCR SV kit (GeneAll, Seoul, South Korea) according to the manufacturer’s instruction and submitted to a commercial laboratory (Axil Scientific Pte Ltd, Singapore) for Sanger sequencing to confirm the 16S rRNA gene target. Raw reads were assembled using DNASTAR™ Lasergene v. 15 (DNASTAR, WI, USA). Sequence matches and species confirmation were analyzed using Basic Local Alignment Search Tool (BLAST) against the rRNA database that is accessible from the National Center for Biotechnology Information website [Citation22].
Enrichment broth to screen presence of ESBL-EC
To further screen for ESBL-EC, an enrichment step was performed by culturing presumptive E. coli-positive colonies in Luria-Bertani (LB; VWR, PA, USA) broth supplemented with 4 mg/l or 8 mg/l of ceftriaxone for 18–20 h at 37 °C (). Ceftriaxone was selected since the MIC threshold of 8 mg/l improves the sensitivity of detecting ESBL-EC [Citation23]. To screen for the presence of ESBL-EC post-challenge with ceftriaxone, 100 μl of each bacterial suspension was spread-plated onto Levine’s eosin methylene blue (L-EMB; Acumedia-Neogen, MI, USA) agar and incubated for 18–20 h at 37 °C. Colonies with a metallic green sheen were further screened for the presence of the yaiO gene using PCR. If the yaiO gene was absent, 16s rRNA gene sequencing was performed to confirm the species identification. Sensitivity tests were conducted on isolates that grew on CHROMID ESBL agar to ascertain if the isolates were true ESBL producers based on their ability to grow on this selective media. This was done by performing a double-disk synergy test to confirm ESBL production where susceptibility to third-generation cephalosporins (amoxicillin/clavulanic acid, ceftazidime, cefotaxime and ceftriaxone) were tested and interpreted according to Clinical and Laboratory Standards Institute guidelines (CLSI M100), along with E. coli ATCC 25922 and K. pneumoniae ATCC700603 strains included as negative and positive controls, respectively.
Results
Enumeration of bacteria present in beach water
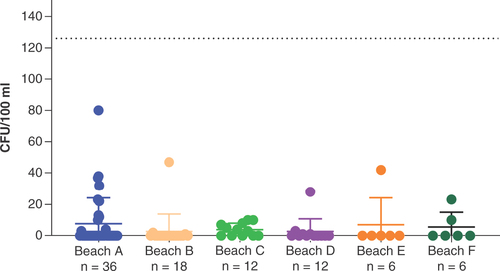
A total of 90 water samples were collected from six different recreational beaches across 3 days for the enumeration of E. coli. E. coli was isolated from 28 samples ( & ). The total E. coli counts ranged from 1 to 80 colony-forming units (CFU)/100 ml of water (average: 4.4 ± 2.1 CFU/100 ml). Among the beaches sampled, Beach C had the highest number of beach water samples with E. coli detected (n = 7; 58%), followed by Beach B (n = 3; 38%), Beach D (n = 4; 33%) and Beach F (n = 2; 33%), Beach A (n = 11; 31%) and Beach E (n = 1; 17%). One site from Beach C had continuous positive E. coli detection in samples collected across the three sampling dates. On the other hand, a few sites at Beach A (n = 3), Beach B (n = 3), Beach D (n = 1) and Beach E (n = 1) yielded no E. coli.
ESBL-EC was not detected in water samples
None of the E. coli isolates were ESBL-EC when screened using the CHROMID® ESBL agar. In contrast, 40 non-E. coli isolates displayed growth on the CHROMID® ESBL agar and were identified via 16S rRNA sequencing to be Pseudomonas spp. (n = 5), Enterobacter spp. (n = 5), Proteus spp. (n = 5), Delftia spp. (n = 4), Marinomonas spp. (n = 1), Raoultella spp. (n = 1) and Alcaligenes spp. (n = 24). Antibiotic susceptibility tests were further carried out on Pseudomonas spp. and Enterobacter spp. isolates. As the isolates demonstrated sensitivity to ceftazidime, a representative of a third-generation cephalosporin, they were thus confirmed to be non-ESBL isolates. Further screening of presumptive E. coli isolates was also carried out using LB broth supplemented with ceftriaxone. Among ceftriaxone-resistant isolates displaying a metallic green sheen on L-EMB, none were ESBL-EC. Instead, 20 were identified to be of the genus Alcaligenes spp. (n = 18) and Providencia spp. (n = 2), respectively.
Discussion
E. coli is one of the most widely used indicators of recent fecal pollution from multiple sources including swimmers, domestic animals or wildlife such as gull waste that may pose a risk to human health [Citation24]. The total E. coli counts were all within the limits of ≤126 CFU/100 ml and ≤500 CFU/100 ml, as stipulated in the US EPA and EU guidelines for recreational beach waters, respectively [Citation25]. Sites with continuous detection of E. coli need to be closely monitored to understand whether there are persistent sources of fecal contamination for effective monitoring and management of coastal water used for recreational activities.
The variable distribution suggests that E. coli may be transiently detected in recreational beach water in Singapore, likely due to varying survivability under environmental conditions. As shown in a previous study, the survivability of E. coli in coastal bathing waters in Spain varied across different locations and was thought to be affected by factors such as water temperature, UV radiation (water turbidity and hours of sunshine), nutrient availability, predation by protozoa and/or salinity [Citation26].
ESBL-EC was not isolated in any of the water samples collected in this study. Previous studies in the Netherlands in 2011–2012 [Citation12], Croatia in 2009–2013 [Citation27] and Norway in 2010 [Citation14] showed the prevalence of ESBL-EC to be 1%, 7.7% and 3.8% in recreational beach waters, respectively. The current findings showed that ESBL-EC was not detected in beach water sampled from popular recreational beaches in Singapore. Of note, ESBL-EC has been detected in previous studies by Zhong et al. and Ng et al. where water samples were collected from a freshwater reservoir and coastal sites used for aquaculture, respectively [Citation17,Citation18]. The presence of ESBL-EC in a freshwater reservoir and coastal seawater aquaculture has also been reported in other countries [Citation28,Citation29]. While the current findings may suggest a low prevalence of ESBL-EC, it should be noted that the study was conducted between the Northeast monsoon and intermonsoon seasons in March 2021. As microbial compositions of surface water in the Singapore Strait may be impacted by seasonal variability [Citation30], a more systematic sample collection at different monsoon seasons will provide a better understanding of the prevalence of ESBL-EC. Indeed, the global Tricycle project recommends environmental samples be collected six to eight times per sampling point [Citation3]. Another limitation of the study was that only a culture-based approach was utilized to determine the presence of ESBL-EC. The inclusion of molecular methods such as quantitative PCR and metagenomics would complement and further support the findings. The collected data will be useful to understand not only their direct impacts on human and animal health but also the potential risk to transfer resistance determinants to other Gram-negative bacteria, to establish guides for mitigation measures.
Conclusion
While no ESBL-EC was detected in all beach water sampled, regular monitoring in different seasons is necessary to understand the prevalence of ESBL-EC and will provide timely risk assessment of exposure to ESBL-EC and contribute to national AMR surveillance.
Extended-spectrum beta-lactamase Escherichia coli (ESBL-EC) has been identified as an indicator organism by the WHO for integrated, multisectoral surveillance of antimicrobial resistance.
In the environment, the risk of ESBL-EC colonization can be through exposure to recreational beach water or urban aquatic environments.
The detection of ESBL-EC in water bodies highlights the potential risk of ESBL-EC exposure to humans. Hence, it is important to survey the prevalence of ESBL-EC in recreational beach waters, where human contact with water bodies is most likely to occur.
The aims of this study were to explore and understand the prevalence of E. coli and ESBL-EC in recreational beaches in Singapore.
In this study, no ESBL-EC was isolated from the collected water samples based on direct plating on CHROMID ESBL agar and pre-enrichment with ceftriaxone prior to plating on CHROMID ESBL agar.
However, a more systematic sample collection across different periods in a year would provide a better understanding of the relationship between seasonality and ESBL-EC trends.
Financial & competing interest disclosure
The study was supported by the Singapore’s Reinvestment Fund: Integrated Programme to Combat Antimicrobial Resistance in the Environment Sector. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Additional information
Funding
References
- Mcdonald KL , GarlandS , CarsonCAet al. Measures used to assess the burden of ESBL-producing Escherichia coli infections in humans: a scoping review. JAC Antimicrob. Resist.3, dlaa104 (2021).
- Puspandari N , SunarnoS , FebriantiTet al. Extended spectrum beta-lactamase-producing Escherichia coli surveillance in the human, food chain, and environment sectors: Tricycle project (pilot) in Indonesia. One Health13, 100331 (2021).
- WHO integrated global surveillance on ESBL-producing E. coli using a “One Health” approach: implementation and opportunities (2022). www.who.int/publications/i/item/who-integrated-global-surveillance-on-esbl-producing-e.-coli-using-a-one-health-approach
- Chereau F , OpatowskiL , TourdjmanM , VongS. Risk assessment for antibiotic resistance in South East Asia. BMJ358, j3393 (2017).
- Karlowsky JA , KazmierczakKM , YoungK , MotylMR , SahmDF. In vitro activity of ceftolozane/tazobactam against phenotypically defined extended-spectrum β-lactamase (ESBL)-positive isolates of Escherichia coli and Klebsiella pneumoniae isolated from hospitalized patients (SMART 2016). Diagn. Microbiol. Infect. Dis.96, 114925 (2020).
- Karlowsky JA , LobSH , DerykeCAet al. Prevalence of ESBL non-CRE Escherichia coli and Klebsiella pneumoniae among clinical isolates collected by the SMART global surveillance programme from 2015 to 2019. Int. J. Antimicrob. Agents59, 106535 (2022).
- Karanika S , KarantanosT , ArvanitisM , GrigorasC , MylonakisE. Fecal colonization with extended-spectrum beta-lactamase-producing enterobacteriaceae and risk factors among healthy individuals: a systematic review and metaanalysis. Clin. Infect. Dis.63(3), 310–318 (2016).
- Tan TY , HsuLY , KohTHet al. Antibiotic resistance in Gram-negative bacilli: a Singapore perspective. Ann. Acad. Med. Singap.37, 819 (2008).
- Young BE , LyeDC , KrishnanP , ChanSP , LeoYS. A prospective observational study of the prevalence and risk factors for colonization by antibiotic resistant bacteria in patients at admission to hospital in Singapore. BMC Infect. Dis.14, 298 (2014).
- Mo Y , SeahI , LyePSPet al. Relating knowledge, attitude and practice of antibiotic use to extended-spectrum beta-lactamase-producing Enterobacteriaceae carriage: results of a cross-sectional community survey. BMJ Open9, e023859 (2019).
- Ding Y , SawW-Y , TanLWLet al. Extended-spectrum β-lactamase-producing and mcr-1-positive Escherichia coli from the gut microbiota of healthy Singaporeans. Appl. Environ. Microbiol.87, e0048821 (2021).
- Blaak H , KruijfPD , HamidjajaRA , HoekAHaMV , HusmanAMDR , SchetsFM. Prevalence and characteristics of ESBL-producing E. coli in Dutch recreational waters influenced by wastewater treatment plants. Vet. Microbiol.171, 448 (2014).
- Hu Y , MatsuiY , RileyLW. Risk factors for fecal carriage of drug-resistant Escherichia coli: a systematic review and meta-analysis. Antimicrob. Resist. Infect. Control11, 31 (2020).
- Jørgensen SB , SøraasAV , ArnesenLS , LeegaardTM , SundsfjordA , JenumPA. A comparison of extended spectrum β-lactamase producing Escherichia coli from clinical, recreational water and wastewater samples associated in time and location. PLOS ONE12, e0186576 (2017).
- Blaak H , LynchG , ItaliaanderR , HamidjajaRA , SchetsFM , HusmanAMDR. Multidrug-resistant and extended spectrum beta-lactamase-producing Escherichia coli in Dutch surface water and wastewater. PLOS ONE10, e0127752 (2015).
- Dorado-García A , SmidJH , PeltWVet al. Molecular relatedness of ESBL/AmpC-producing Escherichia coli from humans, animals, food and the environment: a pooled analysis. J. Antimicrob. Chemother.73, 339 (2018).
- Zhong Y , GuoS , SeowKLG , MingGOH , SchlundtJ. Characterization of extended-spectrum beta-lactamase-producing Escherichia coli isolates from Jurong Lake, Singapore with whole-genome-sequencing. Int. J. Environ. Res. Public Health18, 937 (2021).
- Ng C , ChenH , GohSGet al. Microbial water quality and the detection of multidrug resistant E. coli and antibiotic resistance genes in aquaculture sites of Singapore. Mar. Pollut. Bull135, 475 (2018).
- National Environmental Agengy (2022). www.nea.gov.sg/our-services/pollution-control/water-quality/recreational-beaches
- Molina F , López-AcedoE , TablaR , RoaI , GómezA , RebolloJE. Improved detection of Escherichia coli and coliform bacteria by multiplex PCR. BMC Biotechnol.15, 48 (2015).
- Lane DJ , PaceB , OlsenGJ , StahlDA , SoginML , PaceNR. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl Acad. Sci. USA82, 6955 (1985).
- Altschul S , GishW , MillerW , MyersE , LipmanD. Basic local alignment search tool. J. Mol. Biol.215, 403–410 (1990).
- Huang Y , CarrollKC , CosgroveSE , TammaPD. Determining the optimal ceftriaxone MIC for triggering extended-spectrum β-lactamase confirmatory testing. J. Clin. Microbiol.52, 2228 (2014).
- Korajkic A , McminnBR , HarwoodVJ. Relationships between microbial indicators and pathogens in recreational water settings. Int. J. Environ. Res. Public Health15, 2842 (2018).
- Leonard M , EatonC. Recreational Water Quality Guidelines Update (2021). https://www.esr.cri.nz/assets/WATER-CONTENT/files/Recreational-Water-Quality-Guidelines-Update-September-2021.pdf
- Aragonés L , LópezI , PalazónA , López-ÚbedaR , GarcíaC. Evaluation of the quality of coastal bathing waters in Spain through fecal bacteria Escherichia coli and Enterococcus. Sci. Total Environ.566-567, 288–297 (2016).
- Maravić A , SkočibušićM , CvjetanS , ŠamanićI , FredotovićŽ , PuizinaJ. Prevalence and diversity of extended-spectrum-β-lactamase-producing Enterobacteriaceae from marine beach waters. Mar. Pollut. Bull90, 60 (2015).
- Jeamsripong S , ThaotumpitakV , AnuntawirunS , RoongrojmongkhonN , AtwillER , HinthongW. Molecular epidemiology of antimicrobial resistance and virulence profiles of Escherichia coli, Salmonella spp., and Vibrio spp. isolated from coastal seawater for aquaculture. Antibiotics11, 1688 (2022).
- Rybak B , WawrzyniakN , WolskaL , PotrykusM. Escherichia coli and Serratia fonticola ESBLs as a potential source of antibiotics resistance dissemination in the Tricity water reservoirs. Acta Biochim. Pol.68, 437 (2021).
- Chénard C , WijayaW , VaulotDet al. Temporal and spatial dynamics of bacteria, archaea and protists in equatorial coastal waters. Sci. Rep.9(1), 16390 (2019).