Abstract
Heterogeneity in breast cancer leads to diverse morphological features and different clinical outcomes. There are inherent differences in breast cancer between the populations in Asia and in western countries. The use of immune-based treatment in breast cancer is currently in the developmental stage. The VGH-TAYLOR study is designed to understand the genetic profiling of different subtypes of breast cancer in Taiwan and define the molecular risk factors for breast cancer recurrence. The T-cell receptor repertoire and the potential effects of immunotherapy in breast cancer subjects is evaluated. The favorable biomarkers for early detection of tumor recurrence, diagnosis and prognosis may provide clues for the selection of individualized treatment regimens and improvement in breast cancer therapy.
Lay abstract
We describe the rationale and design for the VGH-TAYLOR study, which includes Taiwanese patients with breast cancer and with a wide spectrum of clinical scenarios covering different breast cancer subtypes and clinical settings, such as the neoadjuvant, adjuvant and metastatic settings. The gene expression profile and genetic mutations of breast cancer subjects with the primary and recurrent tumors are compared. We also explore whether immune-related gene expression and diversity have any impact on response to treatment and survival. This study aims to discover biomarkers of detection of cancer relapse, diagnosis and prognosis that may enable personalized medicine and improvement in breast cancer treatment.
Clinical trial registration: NCT04626440 (ClinicalTrials.gov).
Breast cancer harbors different histopathological and biological features exhibiting distinct clinical outcomes and behaviors. To improve patient management and the overall survival of patients, studies of the early detection of tumor recurrence, diagnosis, treatment and post-treatment care of patients with breast cancer are increasing [Citation1]. In Taiwan, breast cancer is the second leading cause of cancer-related death in women. The annual incidence of breast cancer has continuously increased over the past 2 decades in Taiwan [Citation2,Citation3].
The clinical practices for breast cancer diagnosis include molecular imaging and biochemical markers. Imaging techniques, such as mammography, ultrasound and MRI, could be implemented for monitoring breast cancer progression. CA15-3, carcinoembryonic antigen and circulating cytokeratins are the most commonly used serum biomarkers [Citation4,Citation5].
After a confirmed diagnosis, clinicopathological characteristics are determined, including tumor–node–metastasis (TNM) staging; histological grade; and the status of the estrogen receptor (ER), progesterone receptor (PR) and HER2 as defined by immunohistochemistry (IHC). Several studies have used IHC-based molecular subtyping as surrogate markers for the prediction of patient outcomes and complete response rates (pCR) [Citation6]. Nevertheless, these diagnostic and prognostic markers are associated with various limitations, including inadequate antibody specificity and sensitivity and being somewhat costly [Citation7].
Previous studies have suggested that multigene expression profiling is significantly associated with patient outcomes. Multigene expression profiling serves as a predictive marker for the response to neoadjuvant chemotherapy in patients with ER+ and HER2- breast cancers [Citation8,Citation9]. In addition, liquid biopsies, such as the analysis of circulating cell-free DNA (cfDNA) and circulating tumor cells, could be used for monitoring breast cancer progression and recurrence. Increasing evidence have suggested that cfDNA is more sensitive than CA15-3 and is tested for diagnosis and prognostication of malignancies [Citation10,Citation11]. Detection of mutant cfDNA serves as an inherent biomarker for breast cancer recurrence [Citation12,Citation13].
Introduction to the trial
The VGH-TAYLOR (Veterans General Hospital TAipei – Yung-Ling foundation sinO-canceR) study (ClinicalTrials.gov: NCT04626440) is a comprehensive precision medicine research project focusing on the heterogeneity of Taiwanese breast cancer patients.
Background & rationale
For the purpose of early detection of tumor recurrence and to estimate the risk of breast cancer recurrence more accurately, gene expression profiling tests have been developed for making treatment decisions [Citation14]. A major caveat of these gene expression profiling studies and outcome data is that the populations used for the development and validation are restricted to patients from western countries, which certainly are not entirely reflective of breast cancer patients in Asia [Citation15–18]. In addition, current treatments are unlikely to cure advanced breast cancer; hence, immunotherapy may be a therapeutic strategy for breast cancer [Citation19]. Despite the promising results in other cancer types, data from phase I/II clinical trials have shown disappointing response rates in patients with advanced triple negative breast cancer (TNBC) receiving an immune checkpoint inhibitor [Citation20,Citation21].
Breast cancer has not only been a major public health problem but also a great socioeconomic impact, contributed by relatively expansive new targeted therapies and anticancer therapies as well as considerable high recurrence rate, leading to occupational incapacity and mortality [Citation22–25]. There are limitations of the current tools for diagnosis and prognosis thus establishing a database of Asian-based genetic profiles may improve breast cancer treatment. Accordingly, this study aims to identify potential biomarkers for the early detection of tumor recurrence, prognosis and diagnosis for breast cancer. In addition, to understand the potential effects of immunotherapy in breast cancer subjects, the expression of immune-related genes and T-cell receptor (TCR) diversity will be determined. Our approach employs a standardized next generation sequencing (NGS) panel across all patients rather than different mutation-specific assays for each patient. This approach involved tracking a personalized mutational signature derived from sequencing pretreatment plasma or tumor tissue but obviated the need for unique assays for each patient.
Breast cancer therapy is unique in its multidisciplinary nature and frequently divided into neoadjuvant versus adjuvant approaches for patients with operable breast cancers [Citation4]. In daily practice, patients are informed of the choice of neoadjuvant or adjuvant approaches according to multifactorial considerations (e.g., breast cancer molecular subtypes, tumor staging, age, performance status and other patient-based factors) and can make final decision for each approach. The current VGH-TAYLOR study protocol is a genomic biomarker study correlative to patients' treatments and designed to grouping study subjects according to their treatment scenario.
Methods
Study design
The VGH-TAYLOR study includes a wide spectrum of clinical scenarios covering different breast cancer subtypes and clinical settings such as the neoadjuvant, adjuvant and metastatic settings. By using several NGS-based platforms (e.g., a multigene tumor panel, cfDNA, the expression of immune response genes and the TCR immune repertoire), we will attempt to find biomarkers for Taiwanese patients with breast cancer.
This study was approved by the ethics committee of the Institutional Review Board ([IRB] approval number: 2018-09-007A) of Taipei Veterans General Hospital and was conducted in compliance with the Helsinki Declaration. The principal investigator agrees to provide the IRB/IEC with all appropriate material, including the informed consent document from participants.
This is a study consisting of 3 years of enrollment and approximately 4 years of follow-up after enrollment. The overall study duration is approximately 7 years. It is planned to enroll approximately 2025 subjects over 3 years, including 1875 subjects with breast cancer and 150 archival breast tumor formalin-fixed paraffin-embedded (FFPE) samples from the established biobank (Biobank of Taipei Veterans General Hospital: https://wd.vghtpe.gov.tw/biobank/Index.action) and/or the paired blood samples (if available) from anonymized female subjects with a confirmed diagnosis of primary invasive breast cancer or with recurrent breast tumors. The aims of this study will be achieved by analyzing the genetic profiling from a large cohort of breast cancer subjects using the method of NGS.
Study objectives
The objectives are the following: to conduct comprehensive genetic profiling of subjects with breast cancer; to identify the differences in the genetic profiling of subjects with breast cancer recurrences, to establish the temporal changes in the genetic profiling of breast cancer subjects using circulating tumor DNA (ctDNA), to identify potential biomarkers for the early detection of breast cancer recurrence and predict patient outcomes and to conduct genetic profiling of the immune system in different subtypes of breast cancer.
Subject grouping & assessment plan
Subjects from biobank and recruited patients will be enrolled in this study. illustrates the study flowchart.
Archival samples from the biobank of Taipei Veterans General Hospital.
TMO comprehensive assay, Oncomine Comprehensive Assay v3; TMO cfDNA assay, Oncomine Breast cfDNA Assay; TMO IRR assay, Oncomine Immune Response Research Assay; TMO repertoire assay, Ion AmpliSeq Immune Repertoire Assay Plus-TCR beta.
†Subjects with breast cancer recurrence at screening.
‡Subjects who do not achieve pathological complete response (non-pCR), have breast cancer recurrence, and with paired tumor formalin-fixed paraffin-embedded tissues available.
§Subjects who are currently receiving the first-line treatment for mBC and have PD within 3 months after initiating the first-line treatment for mBC (subjects with rapid PD).
BC: Breast cancer; mBC: Metastatic breast cancer; PD: Progressive disease; TMO: Thermo Fisher Oncomine; WGS: Whole-genome sequencing.
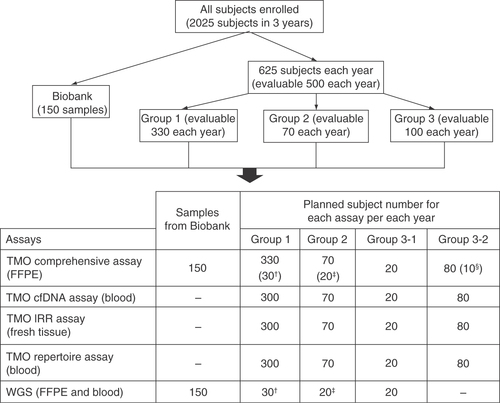
To establish the preliminary baseline genetic profiling of this study, archival FFPE and/or paired blood samples (if blood sample are available) of anonymized female subjects with a confirmed diagnosis of primary invasive breast cancer or with recurrent breast tumors will be assayed ().
Patient enrollment.
After enrollment, individual subject will be assigned into one of the four groups according to the medical management received, the diagnostic stage of breast cancer or the clinical outcome of breast cancer at enrollment. In addition, approximately 625 subjects will be enrolled each year to fulfill the goal of 500 evaluable subjects for genetic profiling analysis, including 330 evaluable subjects in Group 1, 70 evaluable subjects in Group 2, 20 evaluable subjects in Group 3-1 and 80 evaluable subjects in Group 3-2 annually. FFPE tissues, fresh tissues and blood samples will be collected to determine the genetic profiling of breast cancer. The study design of each group is outlined in the following and Supplementary Figure 1.
Group 1 (surgery and adjuvant therapy setting): the evaluable subject number is 330 for each year, including 300 subjects who are planning to undergo surgery as their first-line treatment for breast cancer and 30 subjects with a high risk of recurrence (i.e., with stage III breast cancer, TNBC, or both HER2+ and lymph node-positive [LN+] breast cancer).
Group 2 (neoadjuvant therapy setting): the evaluable subject number is 70 for each year, of subjects who are planning to receive neoadjuvant therapy as the first-line treatment for their breast cancer, including at least 20 subjects who may not achieve a pathological complete response (non-pCR) and may have a breast cancer recurrence.
Group 3 (stage IV breast cancer): the evaluable subject number was 100 for each year, including 20 subjects in Group 3-1 and 80 subjects in Group 3-2.
Group 3-1: subjects diagnosed with de novo and treatment naïve stage IV breast cancer.
Group 3-2: subjects diagnosed with a stage IV breast cancer and with recurrence or stage IV subjects who had received or are currently receiving treatments for breast cancer.
list all the scheduled visits and preplanned sample collection for each NGS testing. We designed this treatment scenario-based protocol with the assumption that participants are fully intended to receive surgery (for Group 1 and Group 2). However, participants are allowed to change their mind for any reasons. We preplanned the collection of samples for each biomarker assays (visits) according to patients' treatment journey (as listed in ). Because of the preplanned nature of the study, there is some flexibility in protocol schedule based on patients' actual course. For subjects who did not receive surgery as planned for whatever the reasons, they can stay on the study protocol as long as they do not fulfill the protocol-defined withdrawn criteria. For example, Group 2 participants would remain at visit 1 and would enter visit 2 only if they received surgery (). Alternatively, these nonsurgery participants will end study whenever they have disease progressed.
Table 1. Schedule of assessments for Group 1 subjects.
Table 2. Schedule of assessments for Group 2 subjects.
Table 3. Schedule of assessments for Group 3-1 subjects.
Table 4. Schedule of assessments for Group 3-2 subjects.
Eligibility criteria
Inclusion criteria
Archival samples from the biobank:
FFPE samples and the paired blood samples (if available) that were collected from female patients with a confirmed diagnosis of primary invasive breast cancer or with a recurrent breast tumor.
Enrolled subjects should meet all of the following criteria for enrollment:
Female subjects aged ≥20 years old.
Subjects with a confirmed diagnosis of primary invasive breast cancer who are planning to receive treatments for breast cancer. However, subjects who have a breast cancer recurrence at screening or stage IV subjects who had received or are currently receiving treatments for breast cancer can also be enrolled.
Eastern Cooperative Oncology Group (ECOG) performance status ≤3.
Life expectancy ≥3 months.
Subject agrees to provide written informed consent.
Exclusion criteria
Subjects will be excluded if they had a primary cancer other than breast cancer within 5 years of screening for inclusion.
Withdrawal criteria
Archival samples from the biobank:
The tumor content of the FFPE sample is lower than the specified percentage according to the standard of the central laboratory.
The FFPE samples failed the DNA/RNA quality check. The criteria of the DNA/RNA quality check will follow the standard of the central laboratory.
Enrolled subjects will be withdrawn if one of the following conditions occurs:
Subject who withdraws consent.
Subject who refuses to provide specimens for evaluation after enrollment.
Subject for which all samples/specimens fail the DNA/RNA quality check. The criteria of the DNA/RNA quality check will follow the standard of the central laboratory.
Subject who does not have sufficient FFPE samples, tissues or blood samples for genetic profiling analysis by principal investigator's discretion.
Subject who does not return to the clinical site for more than 6 months (based on their medical records) will be considered as lost to follow-up. However, whether this subject should be withdrawn will be based on the PI's discretion.
Data collection & follow-up
The data collection period begins when the subjects enroll and ends at the last time point of data collection for subjects in each study group. A case report form (CRF), together with an electronically recorded system (eCRF) are generated to record all the necessary data. These CRF and guidance for eCRF operation have been sent for IRB approval.
For Group 1: for subjects who are planning to undergo surgery as the first-line treatment for breast cancer followed by adjuvant therapy, the last time point of the data collection will be approximately 3 years after the completion of adjuvant therapy. The date of a confirmed diagnosis of breast cancer recurrence after the completion of adjuvant therapy, or the date of death, whichever comes first, is also defined as the last time point of data collection. For subjects with a breast cancer recurrence at screening, the data will be collected from the date of a confirmed diagnosis of breast cancer to the date when the first breast cancer recurrence is confirmed. Group 2: the final date of data collection is 3 years after surgery (mastectomy or breast-conserving surgery), the date of a confirmed diagnosis of breast cancer recurrence after surgery or the date of death, whichever comes first. Group 3-1: the final date of data collection is 3 years after subjects achieve complete response (CR), partial response (PR) or stable disease (SD) for at least 12 months, the date of a confirmed diagnosis of the first PD after initiating first-line treatment for metastatic breast cancer (mBC), or the date of death, whichever comes first. Group 3-2: for nonrapid PD subjects, the last time point of data collection will be approximately 3 years after the subjects achieve a CR, PR or SD for at least 12 months, the date of a confirmed diagnosis of the first PD after enrollment, or the date of death, whichever comes first; or for rapid PD subjects, the last time point of data collection will be the date of the confirmed diagnosis of rapid PD in the 3-year follow-up period or the date of death.
The clinical records including demographics, anthropometrics, ECOG score, menstruation status, family history of cancer, medical history, details of the clinical characteristics of breast cancer at initial presentation, details of surgical management for breast cancer (date and type of surgical management received for breast cancer – simple mastectomy, modified radical mastectomy or breast-conserving surgery), details of medical management for breast cancer (if any), details of first-line treatment for mBC (if any), response to treatment (for subjects in Groups 2, 3-1 and 3-2 only) and details of the clinical outcomes (events of disease progression, recurrence and deaths) of the breast cancer will be recorded in the case report form.
During the follow-up, response to treatments (for example response to neoadjuvant chemotherapy or response to first-line metastatic treatment) is assessed and defined by the Response Evaluation Criteria in Solid Tumors (RECIST, version 1.1) [Citation26,Citation27]. Survival endpoints in this study protocol include disease-free survival (DFS), recurrence-free survival (RFS), progression-free survival (PFS) and overall survival (OS) as defined in literature [Citation28].
Study procedures (NGS tools)
FFPE tissues will be used for the Oncomine Comprehensive Assay v3 (a TMO comprehensive assay) and whole-genome sequencing (WGS) analysis. The TMO comprehensive assay is an NGS-based targeted sequencing assay that enables the detection of relevant single nucleotide variants (SNVs), gene copy number variations (CNVs), gene fusions and indels from 161 unique genes that are designated to help inform drug discovery research and clinical trial research programs [Citation29,Citation30]. Fresh tissues will be used for the Oncomine Immune Response Research Assay (a TMO IRR assay). The TMO IRR assay is for research purpose in current study protocol, and it is also a targeted sequencing gene expression assay that quantitative evaluates the expression of immune genes involved in tumor-immune interactions, such as those associated with leukocyte subsets, antigen presentation and immune checkpoint pathways. Blood samples will be used for the Oncomine Breast cfDNA Assay (a TMO cfDNA assay), the Ion AmpliSeq Immune Repertoire Assay Plus-TCR beta (a TMO repertoire assay) and WGS analysis. The TMO cfDNA assay is designated to detect several breast-cancer-associated biomarkers including AKT1, EGFR, ERBB2, ERBB3, ESR1, FBXW7, KRAS, PIK3CA, SF3B1 and TP53 from cfDNA [Citation31]. Whereas the TMO repertoire assay is a mRNA-based sequencing for complete characterization of CDR1, CDR2 and CDR3 regions of TCR beta [Citation32] and is for research purpose in current study protocol. All of the assays will be conducted following the manufacturer's instructions (Thermo Fisher Scientific, MA, USA), and details of all assays are available at manufacturer's website (https://www.thermofisher.com/). The time schedules of the visits for the participants and the assessments for each group are provided in .
Data analysis
To determine the comprehensive genetic profiling of breast cancer, the frequency of gene mutations identified by WGS and TMO comprehensive assay will be tabulated by the primary and recurrent tumors (if available) of subjects in the Groups 1, 2, 3-1 and 3-2. To determine the differences of comprehensive genetic profiling for recurrence in breast cancer subjects with the primary and recurrent tumors, the potential gene mutations and types of mutation that associated with breast cancer recurrence will be identified by comparing the comprehensive genetic profiling between the paired primary tumor and recurrent tumor of individual subject. To determine the temporal changes in genetic profiling of breast cancer subjects using cfDNA, the frequency of gene mutations identified by TMO cfDNA assay will be tabulated by different study groups at different time points (, Blood collection for TMO cfDNA assay). To determine the temporal changes in the gene expressions involved in tumor–immune interactions in breast cancer subjects, the results of TMO IRR assay will be compared between the paired primary tumor and recurrent tumor in subjects with BC recurrence. To determine the temporal changes in the TCR diversity in breast cancer subjects, the genetic profiling of TCR will be determined using TMO repertoire assay. The variable regions of TCR sequences and CDR3 will be tabulated by different study groups at different time points.
SNVs (reported in the 1000 Genome Project with a minor allele frequency ≥1%) and copy number variants of breast tumor tissues and cfDNA will be detected and analyzed. The potential gene mutations and types of mutation that associated with breast cancer recurrence after neoadjuvant therapy will be identified by comparing the comprehensive genetic profiling between the paired primary tumor and recurrent tumor of individual subject. Survival endpoints including DFS, RFS, PFS and OS curves will be plotted for breast cancer patients with wild-type or mutated gene. TCR repertoire diversity, including evenness, Shannon diversity, TCR convergence, CDR3 length distribution and usage of TCR V(D)J gene segments, in pCR subjects versus non-pCR subjects will be calculated. A receiver operating characteristic curve analysis is used for the prediction of pCR using these TCR features. To determine the altered genes involved in tumor-immune interactions in pCR subjects versus non-pCR subjects, gene with p-values ≤ 0.05 and fold change ≥1.5 will be selected for candidates. The hierarchical clustering analysis and principal component analysis of genes will be performed. The significant impact of genomic and genetic alterations on clinical efficacy and outcome will be evaluated.
Statistical analysis
In general, continuous variables will be summarized as the number of observations, mean, median, standard deviation, minimum, maximum and 95% CIs. Categorical variables will be summarized as counts and percentages. Logistic regression models will be used to determine the potential biomarkers for distinguishing the subjects. Chi-square tests will be used to determine the differences between groups. Clinicopathological characteristics, including cancer stage, histologic grade, lymphovascular invasion, gene expression and gene mutation, which are associated with survival end points (DFS, RFS, PFS and OS) will be used as variables for the univariate and multivariate Cox hazards model and logistic regression analysis. Survival curves of breast cancer subjects will be generated by the Kaplan–Meier method and compared with the log-rank test. All statistical analysis will be performed using the SAS statistical software system. Unless otherwise specified, all statistical assessments will be performed at the significance level of 0.05.
Informed consent process & NGS results availability to participants
The archival delinkage samples from biobank by which the sample donors have given the informed consent at time of donation will be applied from Biobank of Taipei Veterans General Hospital upon IRB approval. For subjects to be enrolled in this study, the informed consent process will be applied. An IRB-approved informed consent form explains how data safety is kept and all the potential implications of participating, including the possible return of ‘incidental’ findings, in easy-to-understand language. In brief, NGS testing data will be interpreted by investigators, and if necessary, a molecular tumor board to evaluate the benefits of returning NGS testing results to participants and patients will be provided information with regard to use of these NGS data, in particular the results of targeted sequencing from multigene panel and cfDNA. Those data that are for research purpose only, such as results from immune response assay and immune repertoire, will not be made available to the participants. All identifiable patient information will be delinked to guarantee confidentiality and security of the information in the genetic material. Only in charging investigator will be able to discuss returning data with individual-responsible participants.
Discussion
The current study has been designed to achieve the following outcomes: understanding the genetic profiles of different subtypes of breast cancer in Taiwan, assessing the efficacy of different treatments for subjects with breast cancer, defining the molecular risk factors and predicting the potential risk of breast cancer recurrence, assessing the immune repertoire and the potential effects of immunotherapy in subjects with breast cancer and developing new strategies for treating patients with TNBC or late stages of breast cancer.
A needle aspiration or solid biopsy followed by imaging techniques is required to confirm the results of mammography and breast ultrasound. Because the incidence of breast cancer has continuously increased, these methods for breast cancer diagnosis are insensitive and are no longer satisfying the medical demands. The available genomic tests, such as Oncotype DX, MammaPrint, Prosigna and EndoPredict, may be used for identifying patients who will benefit from adjuvant chemotherapy for ER+ and HER2- breast cancer [Citation33]. Most of these products focus on Western populations, and the performance of these genomic test panels in Asian populations remains controversial [Citation16]. In this regard, developing an Asian-based genetic profiling database is crucial for Asian populations with breast cancer.
Liquid biopsies are less invasive to access and suitable for serial monitoring cancer progression compared to tissue biopsies. In the bloodstream, the release of cfDNA results from apoptosis, necrosis and secretion by cells [Citation34]. A proof-of-concept analysis indicated that cfDNA carries tumor-specific alterations and there is a significant negative correlation between ctDNA levels and overall survival in mBC [Citation11]. Likewise, ctDNA harbors genetic and epigenetic alterations of tumors, suggesting potential roles as cancer biomarkers. ESR1 and PIK3CA mutations in ctDNA may be linked to drug resistance in hormone receptor-positive breast cancer [Citation35,Citation36]. The US FDA-approved a ctDNA assay, the Cobas EGFR Mutation Test, for use as a companion diagnostic test for metastatic non-small-cell lung cancer eligible for therapy with erlotinib [Citation37].
Immunotherapy has already been used for treating several types of cancer, such as melanoma, lung cancer, acute lymphoblastic leukemia and bladder cancer [Citation38]. Immune checkpoint blockade by antibodies against PD-1/PD-L1 and CTLA-4 exhibits long-lasting antitumor responses in multiple cancers [Citation39]. Although breast cancer is considered to be a poorly immunogenic tumor type, mounting evidence has suggested that the immune system contributes to the prognosis and the chemotherapy response in breast cancer [Citation40]. The tumor infiltrating lymphocytes (TILs) have been reported to be positive prognostic markers, and they are predictive of a therapeutic benefit in patients with breast cancer. High levels of PD-L1 expression has a positive correlation with the presence of TILs in HER2-positive breast cancer and TNBC [Citation41–43]. Recently, atezolizumab, a PD-L1 inhibitor, plus nab-paclitaxel, has been approved for use in patients with PD-L1 positive TNBC [Citation40].
TCR recognizes peptide-MHC epitopes, initiating adaptive immune responses. Activation of the responding T cells results in clonal expansion and their progeny inherit an identical TCR sequence. The generation of a highly diverse TCR repertoire results from random recombination of TCR V(D)J gene segments [Citation44]. The development of NGS-based TCR repertoire analysis provides a powerful tool to study the complexity of the adaptive immune system and cellular immunology. Determination of the TCR repertoire diversity and complementarity-determining regions can be employed to identify biomarkers of immune responses and immune-mediated adverse events [Citation45]. Recent studies have provided evidence that TCR repertoire sequencing can serve as a biomarker of the immune response in cancer patients receiving immunotherapy. TCR sequencing acts as a biomarker for TIL clonal expansion after anti-CTLA-4 antibody ipilimumab therapy in breast cancer [Citation46].
Conclusion
This study is designed as comprehensive precision medical research of Taiwanese patients with breast cancer. The outcomes of this study will provide information about genetic profiles, the efficacy of different treatments including immunotherapy, the risk factors of recurrence and the TCR repertoire in patients with breast cancer. The identification of biomarkers for early detection of breast cancer recurrence and prognosis as well as the prediction of responses to treatments may improve treatments for TNBC and advanced breast cancer.
Executive summary
In Taiwan, breast cancer is the most common cancer and the leading cause of cancer death in women.
Since the incidence of breast cancer has continuously increased, the clinical practices for breast cancer diagnosis are insensitive and are no longer satisfying the medical demands.
Background and rationale
For the purpose of early detection of tumor recurrence and to estimate the risk of breast cancer recurrence more accurately, gene expression profiling tests have been developed for making treatment decisions.
Our approach employed a standardized next generation sequencing panel which involved tracking a personalized mutational signature derived from sequencing pretreatment plasma or tumor tissue.
Study design & eligibility criteria
The VGH-TAYLOR study (ClinicalTrials.gov: NCT04626440) includes a wide spectrum of clinical scenarios covering different breast cancer subtypes and clinical settings such as the neoadjuvant, adjuvant, and metastatic settings.
It is planned to enroll approximately 2025 subjects over 3 years, including 1875 subjects with breast cancer and 150 archival breast tumor formalin-fixed paraffin-embedded samples from the established biobank.
The study population will consist of the Taiwanese patients ≥20 years of age with primary invasive breast cancer who are planning to receive treatments for breast cancer.
Outcome measures/end points
The primary end points are information about genetic profiles, the efficacy of different treatments, the risk factors of recurrence and the T-cell receptor repertoire in patients with breast cancer.
The second end points are disease-free survival, recurrence-free survival, progression-free survival and overall survival.
Conclusion
The identification of biomarkers for early detection of breast cancer recurrence, prognosis and the prediction of responses to treatments may improve treatments for advanced breast cancer.
Author contributions
Conceptualization, methodology, investigation, coordination and conception: LM Tseng and CY Liu. Acquisition of data and revising the work: CY Liu, CC Huang, YF Tsai, TC Chao, YS Lin, CJ Feng, YJ Chen, JH Chiu and CY Hsu. Drafting the article: CY Liu and JL Chen. Study coordinator: PJ Lien. All authors approval of the version to be published and agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethical conduct of research
The whole study protocol was reviewed and approved by Institutional Review Board of Taipei Veterans General Hospital (2018-09-007A). Tumors for immunohistochemical study were collected in accordance with the Declaration of Helsinki and informed consents from sample donors were obtained at time of their donation.
Data availability
Taipei Veterans General Hospital retains the ownership of data, results, reports, findings, and discoveries related to this study. Yong-Lin Healthcare Foundation has the priority authorization. The patient data is unavailable to the public. The findings will be published in journals. The data might be available by requests which need to be approved by both institutions (Taipei Veterans General Hospital and Yong-Lin Healthcare Foundation).
Additional file 1
Download TIFF Image (686.2 KB)Acknowledgments
The authors are grateful to the patients at Taipei Veterans General Hospital, who provided contributions to enable this research project. Research was also supported by Biobank, Taipei Veterans General Hospital.
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.tandfonline.com/doi/suppl/10.2217/fon-2021-0131
Financial & competing interests disclosure
This research is funded by Yong-Lin Healthcare Foundation (SINO-CANCER project) under the clinical study protocol no. QCR18002. The funding body's role was limited to funding with no influence on design, analysis and data interpretation and writing the manuscript. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Additional information
Funding
References
- Dai X , LiT, BaiZet al. Breast cancer intrinsic subtype classification, clinical use and future trends. Am. J. Cancer Res.5(10), 2929–2943 (2015).
- Kuo CN , LiaoYM, KuoLNet al. Cancers in Taiwan: practical insight from epidemiology, treatments, biomarkers, and cost. J. Formos. Med. Assoc.119(12), 1731–1741 (2019).
- Liu FC , LinHT, KuoCF, SeeLCet al. Epidemiology and survival outcome of breast cancer in a nationwide study. Oncotarget.8(10), 16939–16950 (2017).
- Jafari SH , SaadatpourZ, SalmaninejadAet al. Breast cancer diagnosis: Imaging techniques and biochemical markers. J. Cell Physiol.233(7), 5200–5213 (2018).
- Mirabelli P , IncoronatoM. Usefulness of traditional serum biomarkers for management of breast cancer patients. Biomed. Res. Int.2013, 685641 (2013).
- Bhargava R , BeriwalS, DabbsDJet al. Immunohistochemical surrogate markers of breast cancer molecular classes predicts response to neoadjuvant chemotherapy: a single institutional experience with 359 cases. Cancer116(6), 1431–1439 (2010).
- Gown AM . Diagnostic immunohistochemistry: what can go wrong and how to prevent it. Arch. Pathol. Lab. Med.140(9), 893–898 (2016).
- Mazo C , BarronS, MooneyC, GallagherWM. Multi-gene prognostic signatures and prediction of pathological complete response to neoadjuvant chemotherapy in ER-positive, HER2-negative breast cancer patients. Cancers (Basel)12(5) (2020).
- Vallon-Christersson J , HakkinenJ, HegardtCet al. Cross comparison and prognostic assessment of breast cancer multigene signatures in a large population-based contemporary clinical series. Sci. Rep.9(1), 12184 (2019).
- Neumann MHD , BenderS, KrahnT, SchlangeT. ctDNA and CTCs in liquid biopsy – current status and where we need to progress. Comput. Struct. Biotechnol. J.16, 190–195 (2018).
- Dawson SJ , TsuiDW, MurtazaMet al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med.368(13), 1199–1209 (2013).
- Beaver JA , JelovacD, BalukrishnaSet al. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin. Cancer Res.20(10), 2643–2650 (2014).
- Oshiro C , KagaraN, NaoiYet al. PIK3CA mutations in serum DNA are predictive of recurrence in primary breast cancer patients. Breast Cancer Res. Treat.150(2), 299–307 (2015).
- Kittaneh M , MonteroAJ, GluckS. Molecular profiling for breast cancer: a comprehensive review. Biomark Cancer5, 61–70 (2013).
- Shen YC , ChangCJ, HsuC, ChengCC, ChiuCF, ChengAL. Significant difference in the trends of female breast cancer incidence between Taiwanese and Caucasian Americans: implications from age-period-cohort analysis. Cancer Epidemiol. Biomarkers Prev.14(8), 1986–1990 (2005).
- Leong SP , ShenZZ, LiuTJet al. Is breast cancer the same disease in Asian and Western countries? World J. Surg. 34(10), 2308–2324 (2010).
- Kan Z , DingY, KimJet al. Multi-omics profiling of younger Asian breast cancers reveals distinctive molecular signatures. Nat. Commun.9(1), 1725 (2018).
- Pan JW , ZabidiMMA, NgPSet al. The molecular landscape of Asian breast cancers reveals clinically relevant population-specific differences. Nat Commun.11(1), 6433 (2020).
- Basu A , RamamoorthiG, JiaYet al. Immunotherapy in breast cancer: current status and future directions. Adv. Cancer Res.143, 295–349 (2019).
- Gaynor N , CrownJ, CollinsDM. Immune checkpoint inhibitors: key trials and an emerging role in breast cancer. Semin. Cancer Biol.doi:10.1016/j.semcancer.2020.06.016 (2020) ( Epub ahead of print).
- Mina LA , LimS, BahadurSW, FirozAT. Immunotherapy for the treatment of breast cancer: emerging new data. Breast Cancer (Dove Med Press)11, 321–328 (2019).
- Binns C , LowWY, LeeMK. Breast cancer: an increasing public health problem in the Asia Pacific region. Asia Pac. J. Public Health25(5), 364–367 (2013).
- Youlden DR , CrambSM, YipCH, BaadePD. Incidence and mortality of female breast cancer in the Asia-Pacific region. Cancer Biol. Med.11(2), 101–115 (2014).
- Ghoncheh M , MomenimovahedZ, SalehiniyaH. Epidemiology, incidence and mortality of breast cancer in Asia. Asian Pac. J. Cancer Prev.17(S3), 47–52 (2016).
- Ji P , GongY, JinML, HuXet al. The burden and trends of breast cancer from 1990 to 2017 at the global, regional, and national levels: results from the global burden of disease study 2017. Front. Oncol.10, 650 (2020).
- Schwartz LH , LitiereS, DeVries Eet al. RECIST 1.1-Update and clarification: From the RECIST committee. Eur. J. Cancer62, 132–137 (2016).
- Schwartz LH , SeymourL, LitiereSet al. RECIST 1.1 – Standardisation and disease-specific adaptations: Perspectives from the RECIST Working Group. Eur. J. Cancer62, 138–145 (2016).
- Gourgou-Bourgade S , CameronD, PoortmansPet al. Guidelines for time-to-event end point definitions in breast cancer trials: results of the DATECAN initiative (Definition for the Assessment of Time-to-event Endpoints in CANcer trials). Ann Oncol.26(12), 2505–2506 (2015).
- Qu X , YeungC, ColemanIet al. Comparison of four next generation sequencing platforms for fusion detection: Oncomine by ThermoFisher, AmpliSeq by illumina, FusionPlex by ArcherDX, and QIAseq by QIAGEN. Cancer Genet.243, 11–18 (2020).
- Dehghani M , RosenblattKP, LiL, RakhadeM, AmatoRJ. Validation and clinical applications of a comprehensive next generation sequencing system for molecular characterization of solid cancer tissues. Front. Mol. Biosci.6, 82 (2019).
- Nteliopoulos G , PageK, HillsAet al. Comparison of two targeted ultra-deep sequencing technologies for analysis of plasma circulating tumour DNA in endocrine-therapy-resistant breast cancer patients. Breast Cancer Res, Treat.doi:10.1007/s10549-021-06220-9 (2021) ( Epub ahead of print).
- Looney TJ , Topacio-HallD, LowmanGet al. TCR convergence in individuals treated with immune checkpoint inhibition for cancer. Front. Immunol.10, 2985 (2019).
- Duffy MJ , HarbeckN, NapMet al. Clinical use of biomarkers in breast cancer: Updated guidelines from the European Group on Tumor Markers (EGTM). Eur. J. Cancer.75, 284–298 (2017).
- Wan JCM , MassieC, Garcia-CorbachoJet al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer.17(4), 223–238 (2017).
- Fribbens C , O'LearyB, KilburnLet al. Plasma ESR1 mutations and the treatment of estrogen receptor-positive advanced breast cancer. J. Clin. Oncol.34(25), 2961–2968 (2016).
- Baselga J , ImSA, IwataHet al. Buparlisib plus fulvestrant versus placebo plus fulvestrant in postmenopausal, hormone receptor-positive, HER2-negative, advanced breast cancer (BELLE-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol.18(7), 904–916 (2017).
- Oliveira KCS , RamosIB, SilvaJMCet al. Current perspectives on circulating tumor DNA, precision medicine, and personalized clinical management of cancer. Mol. Cancer Res.18(4), 517–528 (2020).
- Kruger S , IlmerM, KoboldSet al. Advances in cancer immunotherapy 2019 – latest trends. J. Exp. Clin. Cancer Res.38(1), 268 (2019).
- Topalian SL , DrakeCG, PardollDM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell.27(4), 450–461 (2015).
- Garcia-Aranda M , RedondoM. Immunotherapy: a challenge of breast cancer treatment. Cancers (Basel)11(12), 1822 (2019).
- Ayoub NM , Al-ShamiKM, YaghanRJ. Immunotherapy for HER2-positive breast cancer: recent advances and combination therapeutic approaches. Breast Cancer (Dove Med Press)11, 53–69 (2019).
- Garcia-Teijido P , CabalML, FernandezIP, PerezYF. Tumor-Infiltrating lymphocytes in triple negative breast cancer: the future of immune targeting. Clin. Med. Insights Oncol.10(Suppl. 1), 31–39 (2016).
- Loi S , MichielsS, SalgadoRet al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: results from the FinHER trial. Ann. Oncol.25(8), 1544–1550 (2014).
- Venturi V , ThomasPG. The expanding role of systems immunology in decoding the T cell receptor repertoire. Curr. Opin. Syst. Biol.12, 37–45 (2018).
- Bradley P , ThomasPG. Using T cell receptor repertoires to understand the principles of adaptive immune recognition. Annu. Rev. Immunol.37, 547–570 (2019).
- Page DB , YuanJ, RedmondDet al. Deep sequencing of T-cell receptor DNA as a biomarker of clonally expanded TILs in breast cancer after Immunotherapy. Cancer Immunol. Res.4(10), 835–844 (2016).
