Abstract
Aim: Expression of EGFP was investigated to ascertain the strength and specificity of CMV, U6 and hepatitis B virus (HBV) core promoters in hepatic and non-hepatic cells. Materials & methods: pSilencer-2.1 plasmid vector is known for siRNA-based inhibition. To achieve target-specific correction of disease-causing genes, pSilencer–GFP was constructed. For liver specific expression of therapeutic genes, endogenous U6 promoter of pSilencer-2.1 was replaced with HBV core promoter and ubiquitously active CMV promoter. Results: Transfection results showed that GFP expression under the control of HBV core promoter was higher in hepatic Hep3B than non-hepatic HEK293T cells. Conclusion: HBV core promoter could lead to specific expression in hepatocytes, which might be used in gene therapy of liver diseases as well as for siRNA-based therapeutic strategies.
Hepatitis B is a deadly liver infection caused by hepatitis B virus (HBV) which can progress further from acute to a chronic life-threatening condition. It is a major health problem globally, which may possibly lead to liver fibrosis, cirrhosis and hepatocellular carcinoma. Not much success has been achieved in complete eradication of hepatitis B with an estimated 296 million chronically infected persons [Citation1,Citation2]. The present-day anti-HBV therapeutics are unable to eradicate the virus from the infected cells. Current therapies for the management of chronic hepatitis B (CHB) are limited to IFN-α or pegylated-IFN-α and any of the five US FDA-approved nucleoside and nucleotide analogues. Despite availability of the vaccine, the coverage rate remains unsatisfactory in high endemic areas [Citation3]. Treatment of patients with present-day drugs is mostly unsatisfactory because of the emergence of antiviral-resistant HBV mutants or adverse side effects, warranting new therapeutic strategy. Viral breakthrough and associated mutations are known among CHB patients receiving tenofovir and/or entecavir. In order to designate with certainty whether these mutations are truly antiviral resistant, more clinical and in vitro studies are needed [Citation4,Citation5]. Emergence of drug resistance is not an exclusive problem of chronic hepatitis B; there are several viral and non-viral human diseases where nucleoside analogue resistance, non response to the available therapeutic regimens, disease break through and relapse of disease occurs. Hence, there has always been a serious need of alternative therapies which could reduce the burden associated with decreasing efficacy and increasing failure of the currently available therapeutic regimens. Therefore, developing new therapeutic measures and investigating the pathogenesis of CHB remains imperative [Citation6,Citation7].
The liver is a key organ and plays a variety of essential roles, the organ is involved in almost all the biochemical pathways and is composed of various forms of cells. Its highly specialized hepatocytes carry out various functions. The hepatitis virus infects the hepatocytes with high efficiency [Citation8]. The liver being the target for hepatitis B viral infection, it is important to improve the efficacy of gene therapy targeting the disease-causing gene. Vectors exploited for gene therapy necessitate the three vital components of an expression cassette, in other words, promoter, therapeutic gene and a polyadenylation signal.
A promoter is essential to control expression of the therapeutic gene. An enhanced and specific expression of therapeutic gene aided with specialized promoter is crucial for effective knockdown of the target gene. Targeting therapeutic genes to the liver with the help of liver-specific promoters can be used to achieve a potent and stable gene expression. Target-specific delivery of the therapeutic gene can be explored by gene therapy [Citation9,Citation10]. Gene therapy offers a great potential for the correction of disease-causing genes by employing the use of viral and non-viral vectors, both of which present benefits and risks. Predominant viral vectors exploited for gene delivery are adenoassociated viral vectors, lentiviral vectors, retroviral vectors and foamy virus vectors. Even though the viral vectors exhibit comparatively higher efficiency of transfecting host cells in comparison to non-viral methods, their considerable drawbacks are their immunogenicity and cytotoxicity. The viral vector integration characteristics, leading to insertional mutagenesis, also contribute to their limitations. Non-viral vectors bear some advantages when compared with the viral vectors: handling is easier, capacity for DNA sequences is very high, low toxicity, low cost, biosafety and they can be specifically targeted to a tissue. Non-viral vectors also have drawn considerable attention due to their lesser immunotoxicity. The key limitations of non-viral vectors are their low transfection efficiency. Non-viral vectors include liposome, DNA protein complexes, naked DNA and plasmid DNA [Citation11–13]. Since plasmid-based vectors have a tendency of target-specific delivery, we used pSilencer-2.1 vector to explore the possibility of liver-specific inhibition of the target gene by replacing its endogenous promoter with hepatitis B viral core promoter. pSilencer-2.1 vector is a siRNA expression vector. It is used for siRNA-based inhibition studies by employing the use of human U6 promoter for its expression. In order to achieve the ultimate goal destined to inhibit the viral life cycle of hepatotropic viruses as well as correction of liver-specific disease-associated genes, the present study has been conducted with the aim of replacing the pSilencer-2.1 vector’s endogenous promoter with hepatitis B viral promoter, followed by evaluation of hepatocytes specificity.
Materials & methods
PCR for construction of pSilencer–GFP expression vector
PCR was employed to amplify the GFP gene from pEGFP vector (Addgene, USA). pEGFP primers with overhangs for restriction enzymes BamHI and HindIII (NEB) were synthesized. To amplify the GFP gene, the upstream primer, 5′ TA GGA TCC ATG GTG AGC AAG GGC GA 3′ and the downstream primer, 5′ CCC AAG CTT TTA CTT GTA CAG CTC GTC C 3′ (IDT, India) were used. The amplicon of GFP gene was ligated into pSilencer-2.1 vector (Thermo Fisher Scientific, MA, USA). The recombinant plasmid was then purified from transformed Escherichia coli DH5α cells, and was subjected to double digestion with BamHI and HindIII enzymes. The resulting construct pSilencer–GFP was confirmed by sequencing with M13F(-40) primer.
Design & construction of chimeric liver-specific & non liver-specific promoters
To determine the relative strength of viral and non-viral promoters, we reconstructed the pSilencer–GFP vector. To enhance the GFP expression in hepatic cells we decided to replace the reporter plasmid (pSilencer–GFP) endogenous U6 promoter with strong liver-specific HBV core promoter. HBV core promoter was amplified from a plasmid pHBV harbouring 1.3x HBV genome. Proof reading enzyme (high fidelity Taq polymerase) was used for all the cloning related experiments. Primers used to amplify the HBV core promoter were synthesized with overhangs for restriction enzymes EcoRI and BamHI (NEB). To amplify the HBV core promoter the upstream and downstream primers used were 5′ AGT GAATTC GAC CGT GTG CAC TTC GC 3′ and 5′ GT GGATCC AAG AGA TGA TTA GG CAG 3′. The amplicon was ligated into linearized pSilencer–GFP vector. The recombinant plasmid was then purified from transformed E. coli DH5α cells, and was subjected to double digestion with EcoRI and BamHI enzymes. The resulting construct pSilencerHBV–GFP was confirmed by sequencing with M13F(-40) primer. To establish a positive control for assessing the strength of viral and non-viral promoters, CMV promoter was selected. With respect to the experiments carried out in the present study, viral promoter refers only to the tissue-specific HBV core promoter. The CMV promoter is also a viral promoter as far its origin is concerned; however, with reference to the tissue specificity, both CMV as well as U6 promoters have throughout been attributed as non-viral and non liver-specific promoters in this study. CMV promoter is a non-viral strong promoter which expresses constitutively in a broad range of cell types. U6 promoter is also a non liver-specific promoter but is weaker than strongly active ubiquitous CMV promoter. To generate a non liver-specific reporter plasmid, the endogenous non-viral U6 promoter was replaced by non-viral CMV promoter. CMV promoter was amplified using pEGFP vector as a template. Primers used to amplify the CMV promoter with EcoRI and BamHI restriction site overhangs were 5′ AAG AAT TCGTGA TGCGGT TTT GGC AGT A 3′ and 5′CAG GAT CCA GCT CTG CTT ATA TAG ACC T 3′. The amplicon was ligated into linearized pSilencer–GFP vector. The recombinant plasmid was then purified from transformed E. coli DH5α cells, and was subjected to double digestion with EcoRI and BamHI enzymes. The resulting construct pSilencerCMV–GFP was confirmed by sequencing with M13F(-40) primer. Diagrammatic representation of the reconstruction of pSilencer-2.1 vector has been shown in . Description of commercially obtained plasmid vectors as well as those engineered in this study has been documented in .
Table 1. Information of plasmids involved in the study.
(A) Structure of pSilencer-2.1 vector. (B) The GFP gene was amplified from pEFGP vector and ligated into pSilencer-2.1 vector, resulting in pSilencer–GFP having endogenous U6 promoter (pSilencerU6–GFP). (C) HBV core promoter was PCR amplified from HBV plasmid and ligated into pSilencer–GFP vector resulting in pSilencerHBV–GFP vector. (D) CMV promoter was PCR amplified from pEGFP vector and ligated into pSilencer–GFP vector resulting in pSilencerCMV–GFP vector.
HBV: Hepatitis B virus.

Maintenance of hepatic Hep3B & non-hepatic HEK293T cell line
Hep3B and HEK293T cells were maintained in complete minimal essential medium (Gibco, MA, USA) and Dulbecco modified Eagle medium (Himedia, Mumbai, India) supplemented with 10% fetal bovine serum (Gibco) and 1% antibiotic-antimycotic solution (Himedia) at 37°C in a humidified incubator supplied with 5% CO2. The cells at 80% confluency were trypsinized and passaged at a split ratio of 1:4 or 1:3 on the basis of experimental needs.
To study the efficiency of liver-specific & non liver-specific promoter in HEK293T & Hep3B cells
HEK293T and Hep3B cells were grown in a 24-well plate and were transfected with pSilencer–GFP constructs with U6, HBV and CMV promoters. For transfection experiments, 1 × 105 cells were plated in 24-well plate. The GFP constructs were transfected with lipofectamine 3000 (Invitrogen, MA, USA) at 70–80% of the cell confluency within 24 h. After 48 h of transfection, fluorescence intensity of the GFP gene under the influence of viral and non-viral promoters was measured using fluorescence microscope.
Quantitative analysis of GFP by ImageJ
ImageJ software has been used in providing quantitative assessment of microscopy data [Citation14]. In this study it was used to measure the mean fluorescence intensity (MFI) of the GFP with the help of images acquired in this study. The difference in expression levels of GFP protein under the influence of viral and non-viral promoters in hepatic and non-hepatic cell lines was evaluated. ImageJ software was exploited in fluorescence quantification of microscopic images.
Statistical analysis
Data were representative of three independent experiments which were performed in triplicates; expressed as mean ± standard deviation. Comparisons between groups were made using Two-way analysis of variance (ANOVA) with Bonferroni post hoc tests used for multiple comparisons. Within the group, comparisons were performed using independent t-test. p-values <0.05 were considered significant. GraphPad Prism Software Inc. version 5 (CA, USA) was utilized for all the statistical analyses.
Results
Construction of reporter plasmid vector containing GFP gene
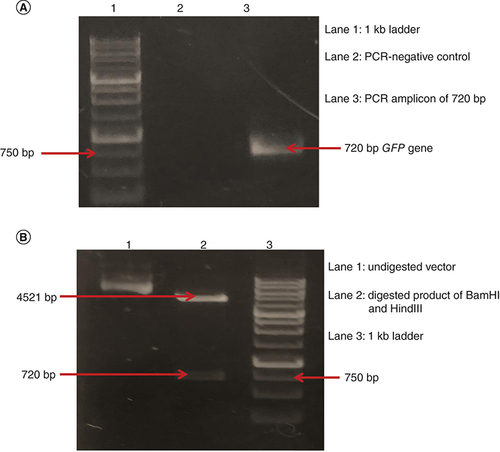
Agarose gel analysis on 1% gel showed that GFP gene of 720 bp was amplified from pEGFP vector by using gene-specific primers (A). After the amplification, the GFP gene was ligated in pSilencer-2.1 vector using T4 DNA ligase. The resulting construct, pSilencer–GFP, was successfully transformed in E. coli DH5α cells. Restriction digestion analysis was done on the isolated pSilencer–GFP construct to confirm the presence of GFP gene. Two separate bands approximately of 4521 bp showing digested vector and 720 bp showing the digested GFP gene were observed (B).
(A) Lane 1: 1 kb DNA ladder. Lane 2: negative control of PCR. Lane 3: 720 bp amplicon of GFP gene. (B) Lane 1: different conformations of pSilencer–GFP construct. Lane 2: BamHI and HindIII digested pSilencer–GFP construct showing digested vector of 4521 bp and digested GFP gene of 720 bp. Lane 3: 1 kb DNA ladder.
Reconstruction of pSilencer–GFP vector
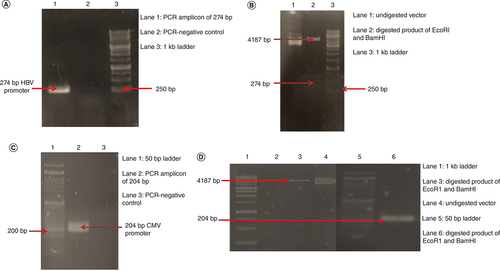
For the reconstruction of the GFP vector HBV core promoter was amplified using pHBV genome as a template. The amplicon was of 274 bp (A). After the amplification, the HBV core promoter was ligated in pSilencer–GFP vector using T4 DNA ligase. Following ligation, the isolated construct was double digested. As a result of digestion by BamHI and EcoRI enzymes, agarose gel analysis shows digested vector of 4187 bp and digested insert of HBV core promoter of 274 bp (B). The sequencing results were found to be consistent with the template (pHBV). pSilencerHBV–GFP was successfully constructed. To generate a non liver-specific reporter plasmid, the CMV promoter from a pEGFP vector was successfully amplified by PCR. The size of the amplicon was 204 bp (C). After ligation the resulting construct pSilencerCMV–GFP was transformed, isolated and confirmed by double digestion and sequencing. The restriction digestion analysis shows a band of 4187 bp depicting the digested vector and a band of 204 bp of CMV promoter depicting the digested insert (D). The sequencing results were found to be consistent with the template (pEGFP).
(A) Lane 1: 274 bp of PCR amplified HBVcore promoter. Lane 2: negative control for PCR. Lane 3: 1 kb DNA ladder. (B) Lane 1: undigested pSilencerHBV–GFP construct. Lane 2: EcoRI and BamHI digested vector of 4187 bp and digested insert of 274 bp. Lane 3: 1 kb DNA ladder. (C) Lane 1: 50 bp DNA ladder. Lane 2: 204 bp amplicon of CMV promoter. Lane 3: negative control of PCR. (D) Lane 1: 1 kb DNA ladder. Lane 3: digested vector by EcoR1 and BamHI of 4187 bp. Lane 4: undigested pSilencerCMV–GFP construct. Lane 5: 50 bp DNA ladder. Lane 6: BamHI and EcoRI digested insert of CMV promoter of 204 bp.
Efficiency of liver-specific & non liver-specific promoter in HEK293T & Hep3B cells
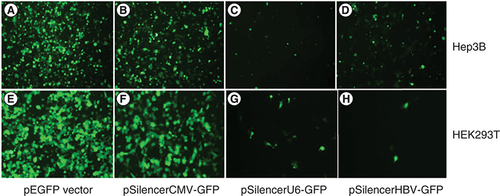
The activities of three different promoters in hepatic and non-hepatic cell lines were studied and compared. Hepatic (HBV) and non-hepatic (CMV and U6) promoters were used to drive the expression of GFP gene in pSilencer–GFP constructs. Among the three different promoters, CMV promoter was found to be the strongest in both Hep3B (A & B) and HEK293T (E & F) cells. Another non liver-specific U6 promoter was found to yield moderate expression levels of GFP gene in Hep3B (C) and HEK293T (G) cells. However, it was observed that the expression of GFP under the control of HBV core promoter was significantly higher in Hep3B as compared with HEK293T cell lines. The liver-specific HBV core promoter exhibited high GFP expression levels in Hep3B cells as compared with HEK293T cells (D). HBV core promoter-mediated GFP expression showed the weakest promoter activity in HEK293T cells (H). As expected, HBV core promoter showed specificity in hepatic cell line. Further, to validate all the images of fluorescence microscopy they were subjected to ImageJ software application (). Readings obtained with ImageJ software were used to plot a graph where mean fluorescence intensity was placed on the Y axis while X axis consisted of relative expression with respect to CMV, U6 and HBV core promoters in hepatic and non-hepatic cells. A prominent difference in expression of GFP in the presence of HBV core promoter is quite evident when compared between HEK293T and Hep3B cells. Hence, the direct visualization of fluorescence microscopy result has been effectively cross verified with the help of ImageJ software application [Citation15].
Different promoters of liver and non liver-specific origin were tested in Hep3B and HEK293T cells. (A & E) GFP expression levels derived from CMV promoter in pEGFP vector in Hep3B and HEK293T cells, respectively. (B & F) GFP expression levels derived from CMV promoter in pSilencer–GFP vector in Hep3B and HEK293T cells, respectively. (C & G) GFP expression levels derived from U6 promoter in pSilencer–GFP vector in Hep3B and HEK293T cells, respectively. (D & H) GFP expression levels derived from HBVcore promoter in pSilencer–GFP vector in Hep3B and HEK293T cells, respectively.
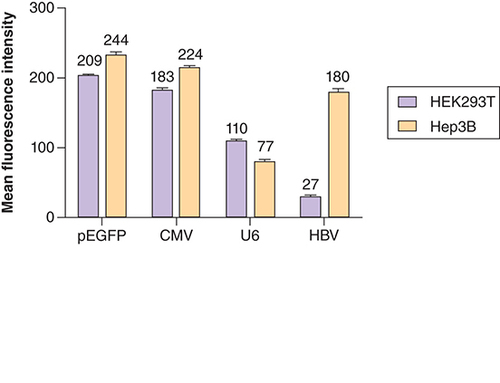
MFI Analysis of difference in GFP expression levels by liver-specific and non liver-specific promoters in Hep3B and HEK293T cells. MFI values for Hep3B cells: pEGFP = 244; pSilencerCMV–GFP = 224; pSilencerU6–GFP = 77; pSilencerHBV–GFP = 180. MFI values for HEK293T cells: pEGFP = 209; pSilencerCMV–GFP = 183; pSilencerU6–GFP = 110; pSilencerHBV–GFP = 27. All the data were indicated as mean ± SD (p < 0.05). Two way ANOVA statistics yielded significant differences between the groups and t-independent test yielded significant differences within the group.
ANOVA: Analysis of variance; HBV: Hepatitus B virus; MFI: Mean fluorescence intensity: SD: Standard deviation.
Discussion
The liver fulfils a great variety of essential functions in the body and is a key organ for most of the metabolic pathways. Liver-targeted gene therapy can be used to treat a number of diseases including primary liver diseases such as hepatitis B. Our goal was to construct a chimera of plasmid vectors which would enable us to discover a highly active liver-specific promoter that can further be used in liver-specific gene therapy. Liver-specific gene targeting is crucial to improve gene therapy of hepatic diseases. One of the many drawbacks of gene therapy is sustained expression of the transgene. To overcome the drawback we explored the idea of cloning a reporter gene in a vector commonly meant for siRNA-based inhibition studies. The pSilencer-2.1 vector was exploited for this purpose [Citation16,Citation17].
In our study, we took a conventional GFP vector (pEGFP-N1 FLAG) that has an endogenous CMV promoter driving the expression of the GFP gene as our reference vector. On the basis of the ubiquitously active CMV promoter in pEGFP vector, our data allowed us to categorize the newly synthesized pSilencer–GFP constructs into three groups: active, which was capable of driving the expression level of the GFP gene similar to that of pEGFP vector; moderately active, which was capable of driving the expression of GFP levels nearly three-fold lesser than pEGFP vector in Hep3B cells (; MFI 244 vs 77) and nearly 1.9-fold lesser in HEK293T cells (; MFI 209 vs 110); weakly active, which was able to drive the least expression levels of the GFP gene as compared with the CMV promoter in pEGFP vector. pSilencer-2.1 vector features a human U6 promoter. To accomplish our aim of analyzing the expression activity of liver-specific and non liver-specific promoters in different cell lines, first the vector was engineered to clone a reporter GFP gene. Then, the newly cloned pSilencer–GFP vector was reconstructed to replace its endogenous U6 promoter with a stronger and widely used non liver-specific CMV promoter. The endogenous U6 promoter was also replaced by liver-specific HBV core promoter. Cell line of hepatic origin Hep3B and non-hepatic origin HEK293T cells were used as model system to assess the difference in expression of the GFP gene by different promoters. In an attempt to construct a highly active liver-specific vector, we transfected hepatic Hep3B cells with pEGFP, pSilencerCMV–GFP, pSilencerU6–GFP and pSilencerHBV–GFPconstructs (A–D). As expected, liver-specific pSilencerHBV–GFP construct resulted in a stronger expression of GFP in Hep3B cell line than pSilencerU6–GFP (C vs D). pEGFP and pSilencerCMV–GFP vector resulted in highest expression of the GFP gene but its expression was non specific. The results were in accordance with our expectations as CMV promoter is a strong promoter; it exhibits high level of expression in a variety of cell lines without any specificity. HEK293T cell line was used as non-hepatic cells and was transfected with the above mentioned constructs. For non liver-specific origin the promoter activity measured by GFP expression through fluorescence microscopy was found to be the strongest in CMV promoter. U6 promoter is also a non liver-specific promoter but its expression was found to be lower than CMV promoter but higher than the HBV core promoter [Citation18,Citation19]. Thus, we can conclude that the ability of different promoters to drive the expression of GFP gene was found to be stronger in non liver-specific promoter in non-hepatic HEK293T cells. In our non liver-specific model system, the GFP expression driven by a liver-specific promoter was found to be the weakest (H). It may be concluded that for sustained liver-specific expression of a therapeutic gene HBV core promoter is a suitable candidate (D vs H & ). We can now categorize our promoters with respect to tissue specificity and their respective MFI values as follows for hepatic cell line; active: pEGFP, pSilencerCMV–GFP; moderately active: pSilencerHBV–GFP; weakly active: pSilencerU6–GFP. For non-hepatic cell line the promoters can be categorized as; active: pEGFP, pSilencerCMV–GFP; moderately active: pSilencerU6–GFP; weakly active: pSilencerHBV–GFP.
In light of present observations, pSilencerHBV–GFP cassette could emerge as a promising recombinant vector to target disease-associated transgene in the liver in a substantially specific manner. The disease-associated transgene could be either infecting viral genome or other genes of hepatocytes. Based on the evidence obtained from this study, it could be believed that pSilencerHBV–GFP could suitably be exploited to sustainably inhibit the HBV life cycle taking the siRNA-based therapeutic approaches. Appropriate siRNA or cassettes of siRNA may be used as replacement of GFP in this vector for this purpose. Second, a very promising feature of liver-specific strong expression of GFP exclusively in the hepatocytes (Hep3B in our study; D) under the influence of HBV core promoter and almost negligible expression of the same in non-hepatic cells (HEK293T in this study; H), strongly indicates the advantages associated with tissue-specific therapeutic approaches with minimal possibility of off target inhibition. This finding is in excellent agreement with the readings obtained by ImageJ software ().
Conclusion
Novel chimeric vector harboring HBV core promoter showed nearly seven-fold increase in expression of the reporter GFP gene in a tissue-specific manner, which could possibly be of substantial usefulness in future for the purpose of siRNA-based antiviral approaches against hepatitis viruses and can also be exploited for liver-specific gene therapy.
Future perspective
The novel pSilencerHBV–GFP engineered in this study could be a beneficial reconstruction vector for the purpose of liver-specific gene therapy and also for siRNA-based inhibition of life cycle of hepatotropic viruses, specially the HBV.
GFP gene is successfully expressed in the novel engineered pSilencer vector.
CMV promoter of the novel vector engineered in this study expressed GFP with equal strength in hepatic (Hep3B) and non-hepatic (HEK293T) cells.
U6 promoter of the novel vector engineered in this study expressed GFP also with equal strength but the expression was moderate compared with that with the CMV promoter.
However, contrary to CMV and U6 promoter, hepatitis B virus core promoter of the novel vector engineered in this study did not express GFP with equal strength in hepatic and non-hepatic cells. Rather, it showed stronger efficiency of expression of GFP in hepatic (Hep3B) cells when compared with negligible amount of GFP expression in non-hepatic (HEK293T) cells.
Author contributions
A Anwer and SN Kazim conceived the idea and planned the study. A Anwer conducted the major experiments, analyzed the data and prepared a major part of the manuscript. A Anwer, S Khan, F Amir, M Afroz and SA Azam made contributions to the experiments as well as in critically reviewing the manuscript. SA Bhat, SK Hasan and R Waseem made their contributions with both the experiments and data analysis. S Parveen, A Islam and SN Kazim gave finishing touches to the manuscript.
Financial & competing interests disclosure
Financial support from DST WOS-A grant no. SR/WOS-A/LS-255/2010(G) was granted to M Afroz and DBT project grant no. BT/PR10740/MED/29/79/2008 was granted to SN Kasim. The first author A Anwer acknowledges receipt of the Senior Research Fellowship (SRF), Indian Council of Medical Research (ICMR), Government of India, for the fellowship support under grant no. VIR/Fellowship/34/2019-ECD-I. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- Naggie S , LokAS. New therapeutics for hepatitis B: the road to cure. Annu. Rev. Med., 72, 93–105 (2021).
- World Health Organization . Hepatitis B. http://www.who.int/news-room/fact-sheets/detail/hepatitis-b
- Tang LS , CovertE, WilsonE, KottililS. Chronic hepatitis B infection: a review. JAMA, 319(17), 1802–1813 (2018).
- Suzuki F , SezakiH, HosakaTet al. Virologic analysis of tenofovir resistance in a patient with chronic hepatitis B experiencing viral breakthrough during combination treatment with tenofovir disoproxil fumarate and entecavir. Hepatol. Res., 51(4), 503–508 (2021).
- Jiang D , WangJ, ZhaoXet al. Entecavir resistance mutations rtL180M/T184L/M204V combined with rtA200V lead to tenofovir resistance. Liver Int., 40(1), 83–91 (2020).
- Zoulim F , DurantelD. Antiviral therapies and prospects for a cure of chronic hepatitis B. Cold Spring Harb. Perspect., 5(4), a021501 (2015).
- Petersen J , ThompsonAJ, LevreroM. Aiming for cure in HBV and HDV infection. J. Hepatol., 65(4), 835–848 (2016).
- Kalra A , YetiskulE, WehrleCJ, TumaF. Physiology, Liver. StatPearls Publishing Copyright © 2021, StatPearls Publishing LLC, FL, USA (2021).
- Singh A , WeberC, VarshneyAet al. Characterization of liver specific promoters in a foamy viral vector pMD09. Acta Virol., 63(2), 162–168 (2019).
- Chen E-Q , SongX-Q, WangY-Let al. Construction of a highly-active, liver-specific transcriptional regulatory element through combination of the albumin promoter and α-fetoprotein enhancer. Plasmid, 65(2), 125–131 (2011).
- Lundstrom K . Viral vectors in gene therapy. Diseases, 6(2), 42 (2018).
- Hardee CL , Arévalo-SolizLM, HornsteinBD, ZechiedrichL. Advances in non-viral DNA vectors for gene therapy. Genes, 8(2), 65 (2017).
- Kamimura K , YokooT, AbeH, TeraiS. Gene therapy for liver cancers: current status from basic to clinics. Cancers, 11(12), 1865 (2019).
- Amir F , SiddiquiZI, FarooquiSRet al. Impact of length of replication competent genome of hepatitis B virus over the differential antigenic secretion. J. Cell. Biochem., 120(10), 17858–17871 (2019).
- Du J , MartinSM, LevineMet al. Mechanisms of ascorbate-induced cytotoxicity in pancreatic cancer. Clin. Cancer Res., 16(2), 509–520 (2010).
- Zheng C , BaumBJ. Evaluation of promoters for use in tissue-specific gene delivery. Methods Mol. Biol., 434, 205–219 (2008).
- Zhang X-N , XiongW, WangJ-D, HuY-W, XiangL, YuanZ-H. siRNA-mediated inhibition of HBV replication and expression. World J. Gastroenterol., 10(20), 2967 (2004).
- Xie N , WangX, ZhangQ, LinY, LiangK, LinJ. The specific expression mediated by promoters of hepatitis B virus in hepatocarcinoma cells. Chin. Ger. J. Clin. Oncol., 5(5), 328 (2006).
- Kramer MG , BarajasM, RazquinNet al. In vitro and in vivo comparative study of chimeric liver-specific promoters. Mol. Ther., 7(3), 375–385 (2003).
