Abstract
Aim: To identify genetic variants associated with greater tobacco consumption in a Mexican population. Patients & methods: Daily smokers were classified as light smokers (LS; n = 742), heavy smokers (HS; n = 601) and nonsmokers (NS; n = 606). In the first stage, a genotyping microarray that included 347 SNPs in CHRNA2–CHRNA7/CHRNA10, CHRNB2–CHRNB4 and NRXN1 genes and 37 ancestry-informative markers was used to analyze 707 samples (187 HS, 328 LS and 192 NS). In the second stage, 14 SNPs from stage 1 were validated in the remaining samples (HS, LS and NS; n = 414 in each group) using real-time PCR. To predict the role of the associated SNPs, an in silico analysis was performed. Results: Two SNPs in NRXN1 and two in CHRNA5 were associated with cigarette consumption, while rs10865246/C (NRXN1) was associated with high nicotine addiction. The in silico analysis revealed that rs1882296/T had a high level of homology with Hsa-miR-6740-5p, which encodes a putative miRNA that targets glutamate receptor subunits (GRIA2, GRID2) and GABA receptor subunits (GABRG1, GABRA4, GABRB2), while rs1882296/C had a high level of homology with Hsa-miR-6866-5p, which encodes a different miRNA that targets GRID2 and GABRB2. Conclusion: In a Mexican Mestizo population, greater consumption of cigarettes was influenced by polymorphisms in the NRXN1 and CHRNA5 genes. We proposed new hypotheses regarding the putative roles of miRNAs that influence the GABAergic and glutamatergic pathways in smoking addiction.
Keywords::
Tobacco consumption is a worldwide epidemic. Many countries have reported an increase in consumption rates, particularly in adolescents and women, and a decrease in the age at initiation [Citation1]. Current trends indicate that by the year 2020, more than 10 million people will die from cardiovascular diseases, chronic obstructive pulmonary disease (COPD) and pulmonary cancer caused by tobacco [Citation2–4]. In Mexico, the prevalence of active cigarette consumption is 21.7% in those 12–65 years of age. On average, this population initiates consumption at 20.4 years old and smokes 6.5 cigarettes per day (cpd). A total of 11.4% of active smokers are addicted to cigarettes, and 58.4% of smokers have attempted to stop smoking [Citation5]. However, the success of smoking-cessation therapy, reported as the percentage of abstinence after 1 year, is 35% [Citation6]. The main addictive component of tobacco is nicotine [Citation7], which exerts its effect on the central nervous system by binding to nicotinic acetylcholine receptors (nAChRs) [Citation8], causing changes in the liberation of neurotransmitters in the nucleus accumbens, which affects various cerebral circuits responsible for drug reinforcement [Citation9,Citation10], the constant search for this substance and the change in cerebral plasticity [Citation11]. The main associated phenotype that predicts the degree of nicotine addiction is the quantity of cpd [Citation12]. Heavy smokers (HS; ≥20 cpd) present a greater degree of addiction, greater difficulty stopping smoking, more withdrawal and tolerance symptoms and higher scores on the Fagerström questionnaire compared with light smokers (LS; <10 cpd) [Citation13,Citation14]. Family and identical twin studies have shown that genetic factors contribute to the risk of consuming tobacco [Citation15,Citation16], with heritability estimations between 40% and 70% [Citation17]. Using genome-wide studies, SNPs associated (p < 1E-07) with the quantity of cpd have been identified. These SNPs are principally located in the genes that encode the nAChR subunits CHRNA5–CHRNA3–CHRNB4 and in neurexin 1 (NRXN1) [Citation18]. A group of genes that are highly associated with nicotine addiction encodes nAChRs. The receptor with the greatest affinity is formed by the α4β2 subunits, which can present the stoichiometries (α4)2(β2)3 and (α4)3(β2)2, of which the first one is more sensitive to nicotine and is found in a greater proportion of individuals who are exposed to nicotine for long periods [Citation19,Citation20]. Associations between nAChR SNPs, nicotine addiction and phenotypes related to cigarette consumption have been reported, particularly in the group of coding genes in chromosome 15 (CHRNA5–CHRNA3–CHRNB4). In 2008, Berrettini et al., who studied a European population, suggested that haplotypes were formed by the CHRNA5/CHRNA3 genes that predispose an individual to nicotine addiction [Citation21]. Subsequently, in 2008, Stevens et al. conducted a study that included LS (<5 cpd) and HS (>30 cpd). These researchers found 13 SNPs that were significantly associated with the HS group [Citation22]. Then, in 2009, Saccone et al. described five SNPs with significant associations with smoking in the CHRNA5 gene, 11 SNPs in CHRNA3 and one associated SNP in CHRNB4. These researchers selected individuals with a score from 0 to 1 on the Fagerström questionnaire as the control group, and they selected individuals who scored ≥4 for the nicotine-dependent group [Citation23]. In this Korean population, two SNPs within the same genetic region were reported as having an association with the quantity of cigarettes consumed [Citation24]. However, the majority of these studies were conducted in European and Caucasian populations; therefore, the results of these studies cannot be extrapolated to populations with different degrees of racial mixing. Identifying whether polymorphisms associated with addiction in these genes exist in the racially mixed Mexican Mestizo population will validate the previously reported population findings, thus helping to clarify the contribution of genetic factors to nicotine addiction and to increased cigarette consumption.
HS: Heavy smoker; LS: Light smoker; NS: Nonsmoker; OR: Odds ratio.
Patients & methods
Study participants
An analytical, cross-sectional study was conducted. This study including active smokers who had smoked for ≥10 years (n = 1343) and who were recruited from smoking cessation support clinics of the Department of Smoking and COPD Research of the Instituto Nacional de Enfermedades Respiratorias Ismael Cosio Villegas, Mexico. Subjects who were smokers were classified based on their consumption of cpd as follows: LS (n = 742), subjects who consumed between 1 and 10 cpd and HS (n = 601), subjects who consumed ≥20 cpd. We excluded moderate smokers (11–19 cpd) because we were interested in comparing the extreme phenotypes. Smokers, either with symptoms (cough, phlegm, wheezing and shortness of breath) or without symptoms, were invited to undergo spirometry at no cost during the COPD Day and World Smoke-Free Day campaigns using mass media advertisements. All smokers who attended our smoking cessation program were also invited to undergo spirometry.
Additionally, a comparison group was included; this group was composed of nonsmokers (NS; n = 606) who were clinically healthy volunteers ≥30 years of age and who did not have addictions to alcohol or other illegal drugs.
All participants were Mestizo Mexicans by birth whose parents and grandparents were born in Mexico, who were male or female, who were not biologically related and who did not have a family history of psychiatric diseases or addictions. Subjects without psychiatric illnesses were included and were evaluated by specialized psychologists who applied the Diagnostic and Statistical Manual of Mental Disorders (DSM-IV-TR) criteria.
The participants were invited to participate in the present research study and were informed about the objective of the study. The participants then signed a letter of informed consent and were provided with an assurance-of-personal-data document. Both of these documents were approved by the Research Institute’s Committee of Science and Bioethical Research (B20-08). Each participant was assigned an alphanumeric key with the purpose of assuring confidentiality. A 15-ml volume of peripheral blood was obtained using venipuncture techniques and collected in a tube with EDTA as an anticoagulant for subsequent DNA extraction using a BDtract DNA Isolation Kit (Maxim Biotech, CA, USA). The DNA was quantified by ultraviolet absorption at a 260-nm wavelength using a NanoDrop instrument (Thermo Scientific, DE, USA). Contamination with organic compounds and proteins was determined by establishing the relationship between the 260/240 and 260/280 readings, respectively. The samples were considered free of contaminants when the relationship was found to be between 1.7 and 2.0.
Genotyping
Identification of SNPs associated with nicotine addiction in a Mexican Mestizo population was carried out in two stages. In the first stage, fine mapping of genes coding for the subunits of the nicotinic cholinergic receptors (CHRNA2 to CHRNA7, CHRNA10, CHRNB2 to CHRNB4) and NRXN1 gene was performed. In the second stage, those SNPs identified as having a significant association in the first stage were validated using an independent sample.
Stage 1
Microarray design & SNP selection
A GoldenGate genotyping microarray was used on an Illumina platform (Illumina, Inc., CA, USA). In total, 347 SNPs were selected by fine mapping in eleven candidate genes (CHRNA2 to CHRNA7, CHRNA10, CHRNB2 to CHRNB4 and NRXN1) using Haploview 4.2 software [Citation25] and by using information in articles published between 2007 and 2011. TagSNPs were included in the microarray if they had a minor allele frequency (MAF) in the Mexican Mestizo population of ≥7% according to the Mexican Genome Diversity Project [Citation26,Citation27].
Selection of ancestry-informative markers
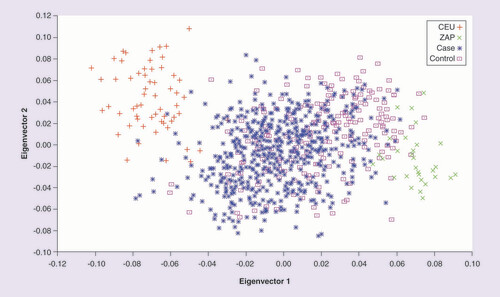
To evaluate the proportion of European and Native American ancestries in individuals participating in our study, we identified 37 ancestry-informative markers (AIMs) that were selected from a panel reported by the Mexican Genome Diversity Project [Citation26,Citation27]. These AIMs were selected using allele frequency differences with delta ≥0.5 for pairwise comparisons between ancestral reference populations, specifically, a European population (CEU-HapMap) and a Native American population (Zapotecas [ZAP] from Oaxaca, Mexico). EIGENSOFT v.4.2 software to calculate fixation index (Fst) statistics for pairwise populations and to obtain the most informative eigenvector [Citation28].
Genotyping & quality control
Genotyping was conducted using the protocol designed by Illumina for the GoldenGate platform (Illumina, Inc., CA, USA) using a Tecan automated liquid handling system (Tecan, Trading AG, Switzerland), which operates under the Illumina protocol. The microarrays were read on a BeadArray Reader (Illumina, Inc.). Genotype acquisition and documentation generation (.ped and .map files) were conducted using GenomeStudio 2011 v.1.0 software (Illumina, Inc.). Subjects who did not meet the call rate criteria (≥95%) were excluded.
Stage 2
During the validation stage, SNPs that were identified as significantly associated with cigarette smoking or nicotine addiction during stage 1 were genotyped. This validation included the thresholds p < 0.001 and odds ratio (OR) ≥ 3.0 for allelic association or genotypic association because the SNPs at this stage required more stringent safeguards [Citation29].
Genotyping was performed using TaqMan allelic discrimination real-time PCR with predesigned probes and a real-time PCR thermocycler (7300 Real-Time PCR Systems, Applied Biosystems, CA, USA). This stage was performed using independent samples with the same characteristics as those samples used in stage 1.
Genotype assignment was performed considering the allelic discrimination and confirmed by absolute quantitation; in addition, three controls without template (contamination controls) were included for each genotyping plate, and 1% of the samples included in the study were genotyped in duplicate as controls for allele assignment. Data interpretation was conducted using Sequence Detection Software (SDS v. 1.4, Applied Biosystems). VIC and FAM fluorophores were used for alleles A and B, respectively.
Statistical analysis
To describe the study population, the statistical program SPSS v.15.0 (SPSS software, IBM, NY, USA) was applied to determine the mean and standard deviation of each variable. To infer the top axes of variation, to estimate the Fst and to correct for stratification in the association analysis, we used a 37 AIM panel to genotype smoker and NS samples and performed a principal components analysis using the smartpca script implemented in EIGENSOFT v.4.2 software [Citation28]. This analysis included the main ancestral population references for Mexican Mestizos, European and Native Americans ancestries, 60 CEU-Hap Map and 30 ZAP reference samples, respectively. Prior to the allelic association analysis, SNPs in the control group (NS) that did not meet Hardy–Weinberg Equilibrium (HWE) and MAF were excluded. The SNPs that complied with the previously established criteria were analyzed with PLINK 1.07 software [Citation30] employing a logistic regression model (with 1 degree of freedom) that included the following factors as covariates: age, sex and the first three principle components obtained from the principal components analysis to correct for ancestry. To identify genetic markers associated with cigarette consumption, the following comparisons were performed: HS versus NS and LS versus NS. The HS versus LS comparison sought to establish genetic markers related to the degree of nicotine addiction.
The significance values obtained in each comparison were corrected by the Bonferroni’s test. The linkage disequilibrium patterns and associated haplotypes were determined using Haploview 4.2 software [Citation25] with the criteria established by Gabriel et al. [Citation31]. For genotypic association analysis, EpiData version 3.1 software was employed [Citation32] using simple 2 × 3 contingency tables, a codominant model and a 95% CI.
In silico analysis & the role of associated SNPs
The SNPs whose associations were validated during stage 2 were evaluated in silico using the Pupasuit 3.1 program [Citation33]. This program can be used to assess the presence of new binding sites for transcription factors and/or the creation or disruption of alternative splicing sites in the gene. To predict potential miRNAs, including the associated SNPs and their potential target genes, the miRDB program was used [Citation34]. We considered only genes (score >80) that are involved in the glutamatergic, GABAergic, serotonergic, dopaminergic and cholinergic pathways, which are the key pathways involved in nicotine addiction. The homology of each predicted miRNA was verified with those reported for humans in miRBase [Citation35], and only those with >80% homology with previously reported miRNAs were considered.
Results
Demographic variables
The demographic variables of the individuals included in both stages are shown in . The mean age and sex significantly differed between the study groups. Because these variables affect cigarette consumption, they were employed as covariates in the genetic association analysis. The HS had been consuming cigarettes for more years and began at earlier ages compared with LS.
Stage 1
Population ancestry analysis
Population stratification was conducted with 37 AIMs (Supplementary Table 1). For this analysis, the participants were grouped as smokers and controls (NS). When the first two most informative eigenvectors were graphed, the study population was observed to be distributed between both reference populations. The genetic distance, expressed as the Fst between the populations of NS and smokers, was 0.030. Differences in the allele frequency existed for the AIMs included in the study compared with the Fst obtained between CEU and ZAP (0.405) (Supplementary Table 2). Consequently, the first three principle components were used as covariates for the association analysis ().
Allele, genotype & haplotype association analyses
In total, 347 SNPs (Supplementary Table 3) were included in the genetic association analysis. After genotyping, two SNPs were excluded for having a genotyping success rate <95%. Prior to the association analysis, 24 of the 345 SNPs were excluded for reporting an MAF <7% and for not complying with HWE in the NS group. Ultimately, 321 SNPs that complied with the previously established quality criteria were employed for the association analysis. This analysis was conducted using a logistic regression model employing ancestry, age and sex as covariates.
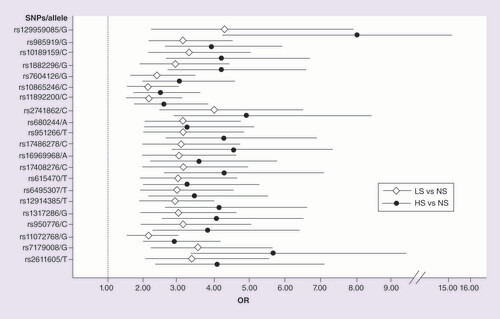
shows the allelic association analysis results obtained for the LS versus NS and HS versus NS comparisons for the nAChR subunits and NRXN1. The calculated p-value was corrected using Bonferroni’s test. Twenty-one SNPs were identified in six genes with statistically significant differences in frequency. NRXN1 and CHRNA5 had the most associated SNPs (7 and 6, respectively). In the comparison between LS versus NS, the p-values were between 7.85E-03 and 5.84E-07, and the ORs were between 2.13 and 4.28. In contrast, in the HS versus NS comparison, the p-values ranged from 8.23E-04 to 1.45E-10 and the ORs were between 2.88 and 7.99. Interestingly, all the SNPs that were found to have an association in the comparison with LS were replicated in the HS versus NS comparison. In addition, the p-values and ORs showed stronger associations. The OR values for each SNP/associated allele in both comparisons are shown in . We did not find associated polymorphisms in the CHRNA2, CHRNA4, CHRNA6, CHRNB2 and CHRNB3 genes.
Additionally, we compared the allele frequencies between smokers with different cigarette consumption cut-offs (HS vs LS). A statistically significant association was obtained (p = 4.26E-03) for the G allele of the NRXN1 SNP rs12995085, which was increased in the HS group (24%) compared with the LS group (13%). This finding suggested that this allele was associated with a greater degree of cigarette consumption in a smoking population (OR: 2.18; 95% CI: 1.53–3.11). Supplementary Table 4 shows the allele data for this comparison.
For the 21 reported SNPs with an allelic association, the genotypes were analyzed using a codominant model. The polymorphisms in the CHRNB4, CHRNA10 and CHRNA7 genes were excluded for having weak OR values (<2.0) compared with SNPs in the NRXN1, CHRNA3 and CHRNA5 genes. The genotype associations for the HS and LS versus NS comparison are shown in . In the HS versus NS comparison, for all the SNPs, the association of the homozygotic genotype for the risk allele was greater than that of the heterozygotic genotype. In fact, in five of the seven associated SNPs, the homozygotic genotype for the risk allele had an OR value that was almost double that of the heterozygotic genotype. A similar trend occurred in the LS versus NS comparison. Again, in the majority of associated polymorphisms, the homozygotic genotype for the risk variant presented a doubled risk compared with its heterozygous genotypes.
The HS versus LS genotype analysis showed statistically significant results (p < 0.001) for the rs12995085 of NRXN1, where the heterozygous genotype (AG) had a frequency of 38.5% in HS and of 25.3% in LS. This factor was associated with the risk (OR: 2.00) of being a HS, and this risk was increased almost four-fold (OR: 7.68) when the homozygotic genotype (GG) was analyzed. This genotype had a frequency of 5.34% in the HS group, whereas it had a frequency of 0.91% in the LS group (Supplementary Table 4).
The haplotype analysis was conducted for all the SNPs in the 11 genes. The linkage disequilibrium was not high in the genetic regions where the CHRNA3, CHRNA5, CHRNB4 and NRXN1 genes are situated (r2 < 0.8). We did not find haplotypes associated with the consumption of tobacco in the three comparisons conducted. The haplotype generated in CHRNA5 is presented in Supplementary Figure 1.
Stage 2
Polymorphisms with values of p < 0.001 and OR ≥ 3.0 in the allelic association analysis (HS vs NS or LS vs NS) or in the genotypic association analysis were chosen. Five SNPs were genotyped in the NRXN1 gene (rs12995085, rs985919, rs10189159, rs1882296 and rs10865246); six, in CHRNA5 (rs680244, rs951266, rs17486278, rs16969968, rs17408276 and rs615470); and three, in CHRNA3 (rs6495607, rs12914385 and rs1317286).
Allelic, genotypic & haplotype associations
Of the 14 SNPs included in the validation, rs12995082 was excluded from the analysis (MAF <5%) in the NS group. The allelic association analysis of the remaining 13 SNPs to compare HS versus NS, LS versus NS and HS versus LS was performed using a logistic regression model with age and sex as covariates. shows the results of four SNPs with statistically significant associations in the comparisons of LS versus NS and HS versus NS. The C allele of rs10865246, located in the NRXN1 gene, was found to be associated in the comparison of HS versus LS (p = 2.79E-02 after Bonferroni’s correction; OR: 1.38; 95% CI: 1.12–1.70).
Genotype analysis was performed using a codominant model. shows the results for the comparisons of HS versus NS and LS versus NS. For the HS versus NS comparison, the OR value almost doubled for the genotypes in the NRXN1 gene (5.78 and 4.07, respectively), and an association with each of the heterozygous genotypes (rs985919/AC and rs1882296/TC) was found (OR: 2.01 and 2.21, respectively). For rs16969968 in CHRNA5, genotypes carrying the risk allele (GA, AA) also retained the association described in stage 1 of the analysis, but with lower OR values (2.05 and 2.51, respectively). Interestingly, rs17408276/TC retained the association found in the comparison of LS versus NS at the second stage of the analysis (p < 0.01; OR: 1.7), even with a higher OR value (2.07). A similar trend was observed in the LS versus NS comparison: rs985919 and rs1882296 homozygous genotypes for the risk allele (CC in both cases) had higher OR values (3.66 and 3.51, respectively), and variants located in the CHRNA5 gene (rs16969968 and rs17408276) showed a modest association (OR: 1.92 and 1.70, respectively) with the heterozygous genotype for each polymorphism.
For the comparison of HS versus LS, a statistically significant association (p = 0.0005) was found with the homozygous genotype (CC) of rs10865246 in the NRXN1 gene (16% [HS] vs 8.3% [LS]; OR: 2.35; 95% CI: 1.48–3.72). Subsequently, a comparison of the variables associated with cigarette smoking (years of smoking, cpd and age of onset of smoking) was performed according to the genotype of each smoker. This analysis compared individuals with a homozygous AA genotype (common genotype) with individuals with AC + CC genotypes (genotypes with allele C, risk). The only statistically significant difference between these individuals was the cpd; subjects homozygous for the common allele had a median of 10 cpd, while individuals with the risk allele had a median of 20 cpd (p = 0.006).
The 13 SNPs that were used for the allelic and genotypic association analyses were then used to perform haplotype analysis. However, no linkage disequilibrium was observed between these genetic variants, and no associated haplotypes were found.
Prioritization of SNPs based on functional properties and miRNA target prediction
Because there is already evidence in the literature of possible biological roles of the identified associations in CHRNA5, only SNPs located in the NRXN1 gene were analyzed. The effect on alternative splicing sites was analyzed using Pupasuit 3.1, and no consequences were found at this level.
miRBase was used to assess the associated alleles; a sequence of 10 bp upstream and 10 bp downstream from the SNP of interest was entered as a query. When rs10865246 was assessed, no target genes were found in the pathways of interest, and potential miRNAs that were generated (with a common or risk allele) had no homology to any previously reported miRNA in humans. We found that rs985919 (with both alleles, common and risk) is part of a sequence of miRNA that potentially targets the genes GRM5 and GABRA2. However, this sequence is not homologous to any human miRNA reported to date. Finally, two potential miRNAs for rs1882296 were found with different target genes (depending on whether the common or risk allele is on the sequence); both genes possess >80% homology with the human miRNAs reported ().
Discussion
Various studies in twins have indicated a high heritability of nicotine addiction, which varies depending on the population studied [Citation36]. These reports were mainly conducted in Asian, African–American and Caucasian populations. However, no data exist for populations of Latin Americans, who exhibit different degrees of racial mixing. In the present work, we evaluated population ancestry using 37 AIMs, and we found some small differences in the Fst between smokers and NS (Fst = 0.030). Consequently, stage 1 of the study considered population ancestry as a covariate to correct for in the allelic association analysis.
In the Mexican Mestizo population, we found and validated two SNPs in NRXN1 (rs985919 and rs1882296) that were associated with tobacco consumption (LS and HS vs NS). In addition, the C allele of rs10865246 is associated with a high degree of nicotine addiction; its frequency is higher in HS compared with LS (44.4 vs 37.9%). In the genotype analysis, this association was stronger when the homozygous genotype was analyzed (CC, OR: 2.35). Members of this gene family are involved in the process of drug addiction according to results obtained in mouse models that revealed a change in the expression of these genes that was induced upon exposure to drugs [Citation37].
Both polymorphisms associated with increased cigarette consumption (rs985919 and rs1882296) are located in introns (18 and 22, respectively). Using Pupasuit software 3.1, we found that neither of these SNPs are in regions that may affect alternative splicing; however, these SNPs are portions of miRNAs. For rs985919, one miRNA was identified whose target genes (GRM5 and GABRA2) encode subunits of glutamate receptors and GABA; however, there is no difference when the miRNA sequence includes the common or risk allele. The miRNA sequence containing the common allele of rs1882296 has five target genes that encode glutamate receptor subunits (GRIA2, GRID2) and GABA receptor subunits (GABRG1, GABRA4, GABRB2), has high homology to Hsa-miR-6740-5p. When we attempted to change the common allele sequence for the risk allele, the potential target genes were GRID2 and GABRB2; this sequence has high homology with Hsa-miR-6866-5p. This analysis shows that a nucleotide change in rs1882296 may result in a different miRNA, which would silence different genes depending on the allele present.
No target genes for rs10865246, which is associated with nicotine addiction, were found in pathways of interest, and to date, there are no miRNAs that include this polymorphism reported for humans in miRBase [Citation38]. In 2007, Bierut et al. found three SNPs (rs10490162, rs12467557 and rs12623467) in intron 1 of the NRXN1 gene in Caucasian and Afro–American populations; these SNPs are associated with the development of nicotine dependence [Citation39]. However, only one of the three SNPs (rs10490162) presented significant associations in European–American populations [Citation40].
Our validation stage shows that polymorphisms rs16969968 and rs17408276 of CHRNA5 are associated with increased consumption of cigarettes. rs16969968 is the most replicated finding (found in our study and in other studies of individuals with European ancestry). The presence of this polymorphism produces a change in the amino acid sequence from aspartic acid (G allele) to asparagine (A allele) in position 398 of the protein, which causes a change in the charge of the second intracellular loop of the α5 subunit [Citation41]. Minor allele (A) of rs16969968 alters the function of the nicotinic cholinergic receptors. In cell assays, changes in the concentration of intracellular calcium are produced when this polymorphism is present. This result provides a possible mechanism by which the decrease in the nicotinic cholinergic receptor function is associated with a significant risk of nicotine addiction. Namely, subjects with the A allele in CHRNA5 require a greater quantity of nicotine to activate the dopaminergic pathway [Citation42]. The homozygotic variant AA for this SNP is significantly associated (p < 0.001) with the group of smokers who consume ≥20 cpd (OR: 2.51). This change has also been proposed to significantly affect the brain signaling of tobacco-consuming subjects, with changes observed in the ventral striatum, amygdala and hippocampus [Citation43]. In 2012, Janes suggested that smokers who present the A variant of rs16969968 are more prone to having memories associated with cigarette consumption and hypothesized that allele A has an important role in the memory process [Citation44]. In a meta-analysis of 16 studies regarding nicotine addiction with various phenotypes associated with tobacco consumption, rs16969968 (p = 5.57E-72) was again found to be associated with a greater consumption of cpd [Citation18]. In 2011, Grucza incorporated multiple gene interactions with nicotine addiction. For nAChRs, Grucza found a significant association with three SNPs of CHRNA5, of which rs16969968 and rs578776 had already been reported previously in at least two different populations [Citation45]. In the European population, SNPs were found that form haplotypes within the CHRNA5–CHRNA3–CHRNB4 and CHRNB3–CHRNA6 groups of genes associated with a risk of nicotine addiction [Citation46]. However, our data showed only an association of cigarette smoking with rs17408276 in the intron position. In individuals of African–American and Caucasian–European ancestry, haplotypes of rs615470 and rs16969968 were reported; these haplotypes were associated with the highest number of cpd [Citation47]. Although the linkage disequilibrium was not high in our population, in 2010, Hansen reported high linkage disequilibrium in another block (rs11637635–rs17408276–rs578776–rs16969968) within the same chromosomal region. This linkage disequilibrium, which was observed in an African–American population, decreases when the African component increases [Citation48]. This evidence suggests that the population distribution of these genetic variants affects the linkage disequilibrium and may have different roles in cigarette consumption.
Our study is the first to evaluate the contribution of genetic variants in the two principle groups of genes associated with nicotine addiction, nicotinic cholinergic receptors and NRXN1, in a Mestizo population. A recent study discovered that the short isoform of NRXN1 interacts with nAChR α4β2, concentrating on the neuron’s presynaptic terminals [Citation49]. Little is known regarding the mechanism by which the long and short forms of neurexin are regulated to generate hundreds of isoforms, but it is possible that certain SNPs can regulate this process and consequently affect the level of neurexin expression. Our analysis provides new hypotheses about the presence of miRNAs that may affect pathways involved in the process of nicotine addiction. These changes in protein expression or silencing of certain messenger RNAs may alter the functions mediated by the nAChRs. These processes (protein expression regulation and mRNA silencing) may jointly participate in the nicotine addiction process.
Conclusion
In conclusion, the present investigation of a Mexican Mestizo population revealed that smoking and, in particular, a greater consumption of cigarettes are influenced by polymorphisms in the NRXN1 and CHRNA5 genes previously described in other populations, in addition to other variants.
Future perspective
The present manuscript has relevant information that can improve our understanding of the genetic factors associated with smoking. The results of this study contribute to the tobacco-related literature. We identified potential genetic markers in the NRXN1 and CHRNA5 genes that are associated with the consumption of cigarettes and with high addiction to nicotine in a Mexican Mestizo population. Little is known regarding the mechanism by which the long and short forms of neurexin are regulated to generate hundreds of isoforms, but it is possible that certain SNPs can regulate this process and consequently affect the level of neurexin expression. Our study provides new hypotheses about the presence of miRNAs that may affect pathways involved in the process of nicotine addiction. In silico analysis suggested that rs1882296 (NRXN1) encodes two potential miRNAs with different target genes in the glutamatergic and GABAergic pathways. Designing biological trials to support these hypotheses and to clarify the roles of these SNPs in the consumption of cigarettes is necessary.
Table 1. Demographic data of the population included in the study.
Table 2. Stage 1: SNPs and associated alleles in the light smoker versus nonsmoker and heavy smoker versus nonsmoker comparisons.
Table 3. Stage 1: SNPs and associated genotypes in the genes NRXN1, CHRNA3 and CHRNA5.
Table 4. Stage 2: SNPs and associated alleles in the light smoker versus nonsmoker and heavy smoker versus nonsmoker comparisons.
Table 5. Stage 2: SNPs and associated genotypes in genes NRXN1 and CHRNA5.
Table 6. Analysis in silico which the potential miRNAs generated for the rs1882296 of NRXN1.
Aim
Smoking is a worldwide epidemic. Several studies have reported the high heritability of nicotine addiction; however, no data exist for Latin–American populations.
Our results will help to clarify the contribution of genetic factors to nicotine addiction and the increased consumption of cigarettes in Mexican Mestizo populations.
Methods
The present study was conducted with smokers who were classified based on their consumption of cigarettes per day (cpd) as light smokers (1–10 cpd, n = 742) and heavy smokers (≥20 cpd, n = 601) and nonsmokers (n = 606).
In the first stage, a genotyping microarray with 384 SNPs including CHRNA2–CHRNA7, CHRNA10, CHRNB2–CHRNB4 and NRXN1 genes was used to analyze individual samples from smokers and nonsmokers.
In the second stage, validation of the polymorphisms identified as associated in the first step was performed using allelic discrimination.
The genetic association analysis was performed using a logistic regression model that included age, sex and ancestry as covariates. To predict the roles of associated SNPs, an in silico analysis was performed.
Results
The polymorphisms rs985919/C and rs1882296/C (NRXN1) and rs16969968/A and rs17408276/C (CHRNA5) are associated with cigarette consumption.
rs10865246/C (NRXN1) was associated (p = 2.79E-02 after Bonferroni’s correction) with high addiction to nicotine (odds ratio: 1.38; 95% CI: 1.12–1.70).
In silico analysis showed that the sequence of miRNA including rs1882296/T (common allele) has high homology to Hsa-miR-6740-5p, while rs1882296/C (risk allele) has high homology to Hsa-miR-6866-5p. This finding suggests that regulation by miRNA will be different in those subjects carrying the risk allele compared with those with the common allele.
Conclusion
In a Mexican Mestizo population, smoking and, in particular, a greater consumption of cigarettes are influenced by the polymorphisms in the NRXN1 and CHRNA5 genes.
Ethical conduct of research
The participants were invited to participate in the present research study and were informed about the objective of the study. The participants then signed a letter of informed consent and were provided with an assurance-of-personal-data document. Both of these documents were approved by the research institute’s Committee of Science and Bioethical Research (B20-08).
Supplemental Figure 1
Download PNG Image (86.2 KB)Supplemental Table 2
Download MS Word (59.3 KB)Supplementary data
To view the supplementary data that accompany this paper, please visit the journal website at:www.tandfonline.com/doi/full/10.2217/pgs-2016-0020
Financial & competing interests disclosure
This paper constitutes a partial fulfilment of the Graduate Program in Biological Sciences of the National Autonomous University of México (UNAM) of G Pérez-Rubio. G Pérez-Rubio acknowledges the scholarship (number: 225531) and financial support provided by the National Council of Science and Technology (CONACyT) and UNAM The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Additional information
Funding
References
- World Health Organization . WHO report on the world epidemic of smoking ( 2011 ). www.who.int/tobacco/global_report/2011/es/ .
- World Health Organization . World No Tobacco Day ( 2013 ). www.who.int/campaigns/no-tobacco-day/2013/event/es/index.html .
- Center for Disease Control and Prevention . Morbidity and mortality weekly reports (MMWRs). Annual smoking-attributable mortality, years of potential life lost, and productivity losses – United States 1997–2001 ( 2005 ). www.cdc.gov/mmwr/preview/mmwrhtml/mm5425a1.htm .
- Liu BQ , PetoR , ChenZMet al. Emerging tobacco hazards in China: 1. Retrospective proportional mortality study of one million deaths . BMJ317 ( 7170 ), 1411 – 1422 ( 1998 ).
- Department of Health . National survey of addictions 2011: tobacco report ( 2011 ). www.conadic.gob.mx .
- Méndez M , UrdapilletaE , SansoresRH , Lara RivasG , Ramírez-VenegasA , Regalado-PinedaJ . Factores que determinan que un paciente ingrese a un programa para dejar de fumar [Factors that determine whether a patient enters a smoking cessation program] . Rev. Inst. Nal. Enf. Resp. Mex.22 ( 1 ), 7 – 13 ( 2009 ).
- Benowitz NL . Clinical pharmacology of nicotine: implications for understanding, preventing, and treating tobacco addiction . Clin. Pharmacol. Ther.83 ( 4 ), 531 – 541 ( 2008 ).
- Azam L , Winzer-SerhanUH , ChenY , LeslieFM . Expression of neuronal nicotinic acetylcholine receptor subunit mRNAs within midbrain dopamine neurons . J. Comp. Neurol.444 ( 3 ), 260 – 274 ( 2002 ).
- Dani JA , BertrandD . Nicotinic acetylcholine receptors and nicotinic cholinergic mechanisms of the central nervous system . Annu. Rev. Pharmacol. Toxicol.47 , 699 – 729 ( 2007 ).
- Zhang T , ZhangL , LiangY , SiapasAG , ZhouFM , DaniJA . Dopamine signaling differences in the nucleus accumbens and dorsal striatum exploited by nicotine . J. Neurosci.29 ( 13 ), 4035 – 4043 ( 2009 ).
- Gipson CD , ReissnerKJ , KupchikYMet al. Reinstatement of nicotine seeking is mediated by glutamatergic plasticity . Proc. Natl Acad. Sci. USA110 ( 22 ), 9124 – 9129 ( 2013 ).
- Chen LS , BakerTB , GruczaRet al. Dissection of the phenotypic and genotypic associations with nicotinic dependence . Nicotine Tob. Res.14 ( 4 ), 425 – 433 ( 2012 ).
- Killen JD , FortmannSP , TelchMJ , NewmanB . Are heavy smokers different from light smokers? A comparison after 48 hours without cigarettes . JAMA260 ( 11 ), 1581 – 1585 ( 1988 ).
- Saccone NL , CulverhouseRC , Schwantes-AnTHet al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD . PLOS Genet.6 ( 8 ), e1001053 ( 2010 ).
- Bierut LJ , DinwiddieSH , BegleiterHet al. Familial transmission of substance dependence: alcohol, marijuana, cocaine, and habitual smoking: a report from the Collaborative Study on the Genetics of Alcoholism . Arch. Gen. Psychiatry55 ( 11 ), 982 – 988 ( 1998 ).
- Li MD . The genetics of nicotine dependence . Curr. Psychiatry Rep.8 ( 2 ), 158 – 164 ( 2006 ).
- Rose JE , BromsU , KorhonenT , DickDM , KaprioJ . Genetics of smoking behavior . In : Handbook of Behavior Genetics (1st Ed.) . KimYK ( Ed. ). Springer , NY, USA , 411 – 432 ( 2009 ).
- Tobacco and Genetics Consortium . Genome-wide meta-analyses identify multiple loci associated with smoking behavior . Nat. Genet.42 ( 5 ), 441 – 447 ( 2010 ).
- Nelson ME , KuryatovA , ChoiCH , ZhouY , LindstromJ . Alternate stoichiometries of alpha4beta2 nicotinic acetylcholine receptors . Mol. Pharmacol.63 ( 2 ), 332 – 341 ( 2003 ).
- Tapia L , KuryatovA , LindstromJ . Ca2+ permeability of the (alpha4)3(beta2)2 stoichiometry greatly exceeds that of (alpha4)2(beta2)3 human acetylcholine receptors . Mol. Pharmacol.71 ( 3 ), 769 – 776 ( 2007 ).
- Berrettini W , YuanX , TozziFet al. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking . Mol. Psychiatry13 ( 4 ), 368 – 373 ( 2008 ).
- Stevens VL , BierutLJ , TalbotJTet al. Nicotinic receptor gene variants influence susceptibility to heavy smoking . Cancer Epidemiol. Biomarkers Prev.17 ( 12 ), 3517 – 3525 ( 2008 ).
- Saccone NL , WangJC , BreslauNet al. The CHRNA5–CHRNA3–CHRNB4 nicotinic receptor subunit gene cluster affects risk for nicotine dependence in African–Americans and in European–Americans . Cancer Res.69 ( 17 ), 6848 – 6856 ( 2009 ).
- Li MD , YoonD , LeeJYet al. Associations of variants in CHRNA5/A3/B4 gene cluster with smoking behaviors in a Korean population . PLoS ONE5 ( 8 ), e12183 ( 2010 ).
- Barrett JC , FryB , MallerJ , DalyMJ . Haploview: analysis and visualization of LD and haplotype maps . BioInformatics21 ( 2 ), 263 – 265 ( 2005 ).
- Silva-Zolezzi I , Hidalgo-MirandaA , Estrada-GilJet al. Analysis of genomic diversity in Mexican mestizo populations to develop genomic medicine in Mexico . Proc. Natl Acad. Sci. USA106 ( 21 ), 8611 – 8616 ( 2009 ).
- Moreno-Estrada A , GignouxCR , Fernández-LópezJCet al. Human genetics. The genetics of Mexico recapitulates Native American substructure and affects biomedical traits . Science344 ( 6189 ), 1280 – 1285 ( 2014 ).
- Patterson N , PriceAL , ReichD . Population structure and eigenanalysis . PLoS Genet.2 ( 12 ), e190 ( 2006 ).
- Grimes DA , SchulzKF . False alarms and pseudo-epidemics: the limitations of observational epidemiology . Obstet. Gynecol.120 ( 4 ), 920 – 927 ( 2012 ).
- Purcell S , NealeB , Todd-BrownKet al. PLINK: a tool set for whole-genome association and population-based linkage analyses . Am. J. Hum. Genet.81 ( 3 ), 559 – 575 ( 2007 ).
- Gabriel SB , SchaffnerSF , NguyenHet al. The structure of haplotype blocks in the human genome . Science296 ( 5576 ), 2225 – 2229 ( 2002 ).
- Public Health Information Department of the General Public Health Administration of the Health Consulate. (Directorate of Galicia), health analysis and health information systems Unit of the Pan-American Health Organization (OPS-WHO). EPIDAT, program for epidemiological analysis of tabulated data . Version3 , 1 ( 2006 ).
- Conde L , VaquerizasJM , DopazoHet al. PupaSuite: finding functional single nucleotide polymorphisms for large-scale genotyping purposes . Nucleic Acids Res.34 ( Web Server ), W621 – W625 ( 2006 ).
- Wong N , WangX . miRDB: an online resource for microRNA target prediction and functional annotations . Nucleic Acids Res.43 ( Database ), D146 – D152 ( 2015 ).
- Griffiths-Jones S , SainiHK , van DongenS , EnrightAJ . miRBase: tools for microRNA genomics . Nucleic Acids Res.36 ( Database ), D154 – D158 ( 2008 ).
- Lessov-Schlaggar CN , PangZ , SwanGEet al. Heritability of cigarette smoking and alcohol use in Chinese male twins: the Qingdao twin registry . Int. J. Epidemiol.35 ( 5 ), 1278 – 1285 ( 2006 ).
- Kelai S , MaussionG , NobleFet al. Nrxn3 upregulation in the globus pallidus of mice developing cocaine addiction . Neuroreport19 ( 7 ), 751 – 755 ( 2008 ).
- Ladewig E , OkamuraK , FlyntAS , WestholmJO , LaiEC . Discovery of hundreds of mirtrons in mouse and human small RNA data . Genome Res.22 ( 9 ), 1634 – 1645 ( 2012 ).
- Bierut LJ , MaddenPA , BreslauNet al. Novel genes identified in a high-density genome wide association study for nicotine dependence . Hum. Mol. Genet.16 ( 1 ), 24 – 35 ( 2007 ).
- Nussbaum J , XuQ , PayneTJet al. Significant association of the neurexin-1 gene (NRXN1) with nicotine dependence in European– and African–American smokers . Hum. Mol. Genet.17 ( 11 ), 1569 – 1577 ( 2008 ).
- Saccone SF , HinrichsAL , SacconeLet al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs . Hum. Mol. Genet.16 ( 1 ), 36 – 49 ( 2007 ).
- Bierut LJ , StitzelJA , WangJCet al. Variants in nicotinic receptors and risk for nicotine dependence . Am. J. Psychiatry165 ( 9 ), 1163 – 1171 ( 2008 ).
- Lips EH , GaborieauV , McKayJDet al. Association between a 15q25 gene variant, smoking quantity and tobacco-related cancers among 17 000 individuals . Int. J. Epidemiol.39 ( 2 ), 563 – 577 ( 2010 ).
- Janes AC , SmollerJW , DavidSPet al. Association between CHRNA5 genetic variation at rs16969968 and brain reactivity to smoking images in nicotine dependent women . Drug Alcohol Depend.120 ( 1 ), 7 – 13 ( 2012 ).
- Grucza RA , JohnsonEO , KruegerRFet al. Incorporating age at onset of smoking into genetic models for nicotine dependence: evidence for interaction with multiple genes . Addict Biol.15 ( 3 ), 346 – 357 ( 2010 ).
- Saccone NL , SacconeSF , HinrichsALet al. Multiple distinct risk loci for nicotine dependence identified by dense coverage of the complete family of nicotinic receptor subunit (CHRN) genes . Am. J. Med. Genet. B Neuropsychiatr. Genet.150B ( 4 ), 453 – 466 ( 2009 ).
- Li MD , XuQ , LouXY , PayneTJ , NiuT , MaJZ . Association and interaction analysis of variants in CHRNA5/CHRNA3/CHRNB4 gene cluster with nicotine dependence in African and European Americans . Am. J. Med. Genet. B Neuropsychiatr. Genet.153B ( 3 ), 745 – 756 ( 2010 ).
- Hansen HM , XiaoY , RiceTet al. Fine mapping of chromosome 15q25.1 lung cancer susceptibility in African–Americans . Hum. Mol. Genet.19 ( 18 ), 3652 – 3661 ( 2010 ).
- Cheng SB , AmiciSA , RenXQet al. Presynaptic targeting of alpha4beta 2 nicotinic acetylcholine receptors is regulated by neurexin-1beta . J. Biol. Chem.284 ( 35 ), 23251 – 23259 ( 2009 ).