Abstract
Aim: Therapy with low-dose amitriptyline is commonly used to treat painful diabetic peripheral neuropathy. There is a knowledge gap, however, regarding the role of variable CYP2D6-mediated drug metabolism and side effects (SEs). We aimed to generate pilot data to demonstrate that SEs are more frequent in patients with variant CYP2D6 alleles. Method: To that end, 31 randomly recruited participants were treated with low-dose amitriptyline for painful diabetic peripheral neuropathy and their CYP2D6 gene sequenced. Results: Patients with predicted normal or ultra-rapid metabolizer phenotypes presented with less SEs compared with individuals with decreased CYP2D6 activity. Conclusion: Hence, CYP2D6 genotype contributes to treatment outcome and may be useful for guiding drug therapy. Future investigations in a larger patient population are planned to support these preliminary findings.
Diabetes is a public health problem worldwide and will be the seventh leading cause of death by 2030 (WHO). Its occurrence is greatest in low- and middle-income countries. In the sub-Saharan African region, almost 7 million people live with diabetes and this number is likely to double by 2025 [Citation1]. Diabetic Peripheral Neuropathy (DPN) is the most common complication of diabetes and occurs in up to 50% of diabetic patients [Citation2]. DPN is defined as ‘the presence of symptoms and/or signs of peripheral nerve dysfunction in people with diabetes, after exclusion of other causes’ [Citation3]. The common clinical symptoms are allodynia, hyperalgesia and spontaneous pain [Citation4]. About 26% of all diabetic patients suffer from neuropathic pain which is characterized as burning and shooting pain in the lower limbs and feet [Citation5]. This discomfort often impacts directly on productivity, as reported by Gore et al., who observed that 65% of working patients with DPN either missed work or experienced decreased work productivity due to pain [Citation6].
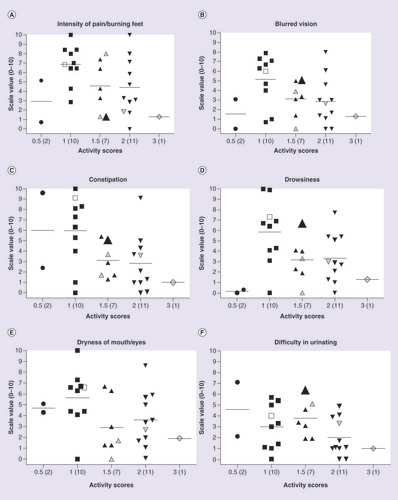
Side effects (SEs) were scored on a scale of 0 to 10 (0 = no SEs and effective treatment; 10 = high number of SEs and ineffective treatment). CYP2D6 activity is represented as activity scores and the number of individuals per category is shown in brackets. Gray-shaded triangles and diamonds are for patients on CYP2C19 inhibitor drugs. The open square denotes a patient on a CYP2D6 inhibitor. The large solid triangle represents patient # 17 carrying the 2509G>T SNP in exon 5 that causes a K245N amino acid change. (B–F) Represent study participants with lower activity scores and severe SEs.
There are no treatments that can restore nerve function, and hence drug therapy is mainly aimed at controlling pain [Citation5]. However, up to two thirds of patients do not obtain adequate pain relief from their current drug regimens [Citation7]. Drug therapy most often includes antidepressants (tricyclic antidepressants [TCAs], selective serotonin reuptake inhibitors and serotonin-noradrenaline reuptake inhibitors), sodium channel blockers and anticonvulsants [Citation8]. Amitriptyline is the drug of choice for treatment of painful peripheral neuropathy in the South African public health sector. Given its cost–effectiveness, it is also widely used in other developing countries [Citation9]. It is a TCA and is administered at doses of 150–250 mg/day for depression. However, when prescribed for pain management, it is used at significantly lower starting doses of 10–25 mg/day [Citation10]. Amitriptyline is mainly metabolized by CYP2C19 and CYP2D6, both of which are members of the CYP450 family of Phase I drug-metabolizing enzymes. The parent drug is rapidly demethylated by CYP2C19 to an active secondary amine, nortriptyline, which has both analgesic and antidepressant effects. Nortriptyline is then hydroxylated by CYP2D6 to form 10 hydroxynortriptyline, an inactive metabolite that is cleared by the kidneys. It has been shown that nortriptyline is predominantly responsible for the adverse drug reactions (ADRs) experienced in amitriptyline therapy [Citation9]. If CYP2D6 clearance of nortriptyline is compromised due to absent or decreased enzyme activity, enhanced receptor occupation could potentially lead to ADRs.
The CYP2D6 gene is located on chromosome 22q13.1 and has nine exons. It is highly polymorphic with over 100 allelic variants and subvariants having been designated by the Human Cytochrome P450 (CYP) Allele Nomenclature Database [Citation11] (many additional variants have been observed, but have not been systematically captured). CYP2D6 represents only 2–4% of hepatic P450 protein, but is responsible for the metabolism of approximately 25% of drugs [Citation12]. CYP2C19 is also highly polymorphic. To date, 35 allelic variants have been defined not counting subvariants. The gene is located on chromosome 10q24 and contributes to the metabolism of about 10% of drugs [Citation13]. The frequencies of CYP2D6 and CYP2C19 genetic variants vary across world populations and ethnic groups [Citation14–16]. Phenotypically, people can be broadly classified according to their CYP2D6 and CYP2C19 activity: poor metabolizers (PMs, no activity), intermediate metabolizers (IMs, decreased activity), normal metabolizers (NMs, wild-type/normal activity) and ultra-rapid metabolizers (UMs, increased activity) [Citation17]. PMs are at a higher risk of exhibiting side effects (SEs) or ADRs, while UMs may have a poor or absent response owing to subtherapeutic drug levels. Some of the SEs for amitriptyline include anticholinergic effects such as constipation, dizziness, dry mouth, blurred vision and urinary retention. The ADRs that are more severe and include cardiotoxicity and CNS toxicity [Citation18].
The Clinical Pharmacogenetics Implementation Consortium (CPIC) has published guidelines providing dose recommendations for TCAs. This includes amitriptyline dosage and drug choice based on a patient’s genotype that is used to optimize drug therapy and thereby minimize ADRs for TCAs according to CYP2D6 and CYP2C19 genotypes [Citation18]. It needs to be noted though, that the recommendations are for the treatment of depressive disorders, and hence are in reference to the higher TCA dose ranges. Since amitriptyline is typically used at lower dosages for the treatment of neuropathic pain, CYP2D6 or CYP2C19 PMs or IMs are less likely to experience ADRs, but rather SEs. The recommendations by CPIC, in other words, that patients on low-dose TCAs should be closely monitored for SEs, have not changed in the recently published guideline update due to lack of information regarding low-dose amitriptyline therapy for neuropathic pain [Citation19]. In our experience with low-dose amitriptyline, we often encounter SEs such as blurred vision, drowsiness, constipation, urinary retention and dryness of mouth/eyes in patients who take amitriptyline at lower dosages, which thus warrants further investigation.
Given the extensive and broad usage of amitriptyline for the management of chronic neuropathic pain in South Africa, and the well-described metabolic disposition of the drug, we hypothesized that pharmacogenetic testing might be useful for tailoring treatment and thereby improving amitriptyline efficacy in these patients. To test this hypothesis, and prior to embarking on a large-scale definitive study, we conducted a pilot investigation with the objectives of demonstrating proof of concept, and generating data for power calculations in order to assist in the planning of a larger scale study. For the purpose of this pilot study, the patient population was restricted to those with painful DPN, and pharmacogenetic testing was limited to CYP2D6, given that CYP2C19 contributes to the ratio of tertiary to secondary amine plasma concentrations; CYP2D6, however, is believed to have a larger impact on overall drug clearance than CYP2C19 [Citation20]. Predicted phenotype status was correlated with SE scores that were derived from a daily questionnaire completed by the study participants.
Methods
Study population & questionnaire
Approval for this observational study was obtained from the University of Pretoria Research Ethics Committee (Ethics approval number 102/2005). Study participants over the age of 18 were recruited at random over a period of 6 months from the Diabetic Clinic at Kalafong Hospital (Atteridgeville, South Africa), and were included in the study after providing written informed consent. Participants were included if they had DPN and were on amitriptyline treatment for the management of chronic pain. Diagnosis of DPN was confirmed using monofilament and vibration sensory tests [Citation21]. A monofilament test is commonly employed to evaluate peripheral nerve sensation in diabetic patients (usually done on the feet). The patient is asked to score the sensations caused by a thin fiber that is used to touch the patient’s foot at various positions. In the vibration sensory test, a vibration fork (128 Hz) is placed on the distal interphalangeal joint where the vibration sense should normally be felt. If it is not felt the vibration is sequentially tested on more proximal bony prominences. The lowest prominence felt is used to describe the level at which vibration can be felt.
Participants received a diary which they completed each day for 7 days following their clinic visit. The diary contained information regarding efficacy and SEs related to amitriptyline. Scoring was based on a rating scale from 1 to 10, where 1 indicated that the participant was completely satisfied and 10 indicated that the participant was completely dissatisfied. The diary comprised six questions that addressed the following for each day: the intensity of pain/burning in feet, blurred vision, constipation, drowsiness, dryness of the mouth and/or eyes, and difficulty urinating. Patients were asked to return the diary to the recruiter or study coordinator on the day of their next clinic visit or another suitable date. For interpretation, average scores of more than 5.0 were regarded as the threshold confirming the presence of pain or one of the recorded SEs.
Information regarding concomitant drugs (including doses) received by the patients, amitriptyline dose and ethnicity was recorded at the time of recruitment.
Sample processing & CYP2D6 characterization
Blood was collected in ethylenediaminetetraacetic acid (EDTA) vacutainers and high-quality genomic DNA isolated from the leukocyte layer using the Maxwell®16 system (Promega, WI, USA). DNA was stored at -20°C prior to analysis.
Amplification of CYP2D6, detection of the CYP2D6*5 gene deletion and gene duplication alleles was achieved using a modified tetramer multiplex assay [Citation22]. Prior to sequencing, amplified products (5.1 kb) of CYP2D6 were treated with FastAP Thermosensitive Alkaline Phosphatase (Fermentas Life Sciences, Vilnius, Lithuania) according to the supplied protocol. Sequencing of the entire gene (introns and exons) including 5′- and 3′-flanking regions (start and end positions for M33388, AY545216 and hg19 coordinates were: -1543–4180, 2658–8381 and 42,528,341–42,522,613, respectively) was performed by Inqaba Biotechnological Industries (Pty) Ltd. (Pretoria, South Africa), using an ABI Big Dye Terminator Cycle Sequencing kit version 3.1 and 3130 XL and 3500 XL DNA analyzer instruments (Applied Biosystems, Inc., CA, USA). Electropherograms were analyzed using CLC DNA Workbench version 5.5 (CLC bio, Aarhus, Denmark) and aligned to the National Center for Biotechnology Information (NCBI) AY545216 reference sequence for the detection and annotation of sequence variations. The assignment of alleles was done according to The Human Cytochrome P450 Allele Nomenclature Database for CYP2D6 [Citation23].
CYP2D6 genotype calls
Alleles were assigned using a set of ‘key’ SNPs defining star (*) alleles as shown in Supplementary Table 1. These assignments were used to determine diplotypes and phenotype prediction as well as to calculate allele frequencies.
The majority of subjects contained additional SNPs that could not or only in part, be reconciled with allele definitions described by the nomenclature database. Novel variant haplotypes were manually inferred by taking into account allele definitions (per the nomenclature database), variants described previously [Citation24] and novel variants found in this study that could unequivocally be determined to be due to the presence of the CYP2D6*5 gene deletion (Supplementary Table 1). A subset of individuals carried novel SNP(s) that could not be assigned to either allele.
CYP2D6 phenotype prediction
CYP2D6 genotypes were translated into phenotype as previously described [Citation25], using the activity score system (Supplementary Table 2). Briefly, the sum of values assigned to each allele provides the activity score that is used to group subjects into the following phenotype groups: 0 indicates PM status; a score of 0.5 indicates IM status; scores between 1 and 2 indicate NM status and a score more than 2 indicates UM status.
Statistical evaluation
The GraphPad Prism version 5 for Windows (GraphPad Software, CA, USA) was used to generate scatter plots.
For power calculations, and given that six measurables were reported in the participant questionnaires, multiple testing was corrected for by applying the Bonferroni inequality principle [Citation26]. Therefore, each variable was measured against a significance level of 0.00833 (i.e., 0.05/6). Based on the data gathered during the preliminary study, the noncentrality parameters of the Analysis of Variance (ANOVA) F-tests [Citation27] were calculated to compare the sample means. Based on the noncentral F-distribution, the power was calculated for overall sample sizes at 30, 60, 90, 120, 150 and 180.
Results
The sample cohort consisted of 21 Black African, nine Caucasian and one Indian participant. Supplementary Table 1 provides a summary of the sequence variations and alleles identified. We observed 42 SNPs, a deletion (del>C), two insertions (ins>G), a four-nucleotide insertion (ins>CAAC) and a series of SNPs defining the intron 1 conversion. We identified seven novel haplotypes: *1 V1, *1 V2, *1 V3, *1 V2xN, *2 V4 and V6 and *2 V1xN, found variants described previously [Citation24] that are not cataloged by the P450 Nomenclature website, but also discovered a number of known and novel SNPs for which haplotype could not be determined. Of the novel SNPs, one was exonic (exon 5, 2509G>T, [K245N]). Supplementary Table 1 provides the genotype assigned to each case considering all SNPs found (column B) and the genotype using only ‘key’ allele-defining SNPs (column D).
CYP2D6 allele frequencies
Allele frequencies (based on genotype assignments shown in Supplementary Table 1 (column D) are summarized in . Considerable differences in frequencies were determined when the data were stratified according to ethnicity. In our Black African patients, the CYP2D6*1 (33%) and *2 (19%) alleles were the most frequent, while the so-called African-specific alleles CYP2D6*17 and *29 were less abundant at frequencies of 12 and 7.0%, respectively. The latter were absent in the Caucasian and Indian patients. The most common alleles among the nine Caucasian patients were CYP2D6*2 and *4 (both 28%) followed by CYP2D6*1 (22%). Gene duplications were only observed in Black African patients. Also notable is the relatively high frequency of the CYP2D6*5 gene deletion (14%) among the Black African patients. The alleles we have observed in our study population are consistent with those previously described for a South African population [Citation28].
Phenotypic prediction of CYP2D6 & correlation with SE profiles
Genotypes were translated into predicted phenotype using the CYP2D6 activity score system [Citation25]. As expected, the majority of study participants (n = 28; 90.32%) were classified as CYP2D6 NMs. Two patients (6.45%) were IM (one Caucasian and one Black African), and one Black African patient had a CYP2D6 genotype predictive of UM status. We did not observe any genotypes predictive of a PM phenotype. For data analysis, subjects were grouped by activity score. As shown in B–F, there is a trend for study participants with lower activity scores to report more severe SEs. These observations are in agreement with the expected higher amitriptyline and nortriptyline plasma levels in participants with decreased CYP2D6 activity.
By assigning average threshold scores of more than 5.0 as confirmation of the presence of a SE, it was possible to qualitatively report on the occurrence of SEs relative to predicted CYP2D6 phenotype (). Using this approach, we observed that IM individuals experienced up to two SEs during the period of evaluation. In contrast, 61% (17/28) of NMs experienced SEs and 39% (11/28) did not experience any SEs. Of those NMs who experienced 1–5 SEs, 53% had an activity score of 1 (less CYP2D6 activity compared with the patients with activity scores of 1.5 or 2). Of note, the UM individual did not report any SEs. The majority of the study participants were on 25 mg of amitriptyline (i.e., 64%; 18/28) at the time of recruitment. Participants were also taking a number of other drugs during the study (5–15 drugs including CYP2D6 or CYP2C19 inhibitors). In order to compare the predicted CYP2D6 phenotypes with participant scores for the severity of SEs, data were grouped into four categories. These include: category 1, no SEs and increased CYP2D6 activity; category 2, no SEs and normal CYP2D6 activity; category 3, SEs and normal CYP2D6 activity; and category 4, SEs and decreased CYP2D6 activity.
Power calculations
Given the distribution of data generated in this pilot study, power calculations were performed to assist in the planning of a larger scale study (Supplementary Table 3). The calculations indicate that for n = 180 subjects, each measurement will have a probability of at least 0.9 of being significant. With a choice of N = 150, one measurement will have a power of slightly below 0.9.
Discussion
Genetic variability of CYP2D6 & phenotype prediction
The most notable novel SNP is 2509G>T that was found in exon 5 in a Black African patient. This nonsynonymous SNP causes an amino acid change (K245N) having the potential to alter enzymatic activity. 2509G>T was observed in a CYP2D6*2/*29 genotype () and was classified as NM for this analysis. This patient experienced all SEs except dryness of eyes and mouth (subject 17 in ) which suggests that K245N may cause decreased CYP2D6 function toward amitriptyline. The variant effect predictor available through Ensemble predicted that K245N has only a moderate impact. Further functional investigations are warranted, however, to fully elucidate the consequences of this amino acid change on catalytic activity.
Although sample size was a limitation in this investigation, allele frequencies () were similar to those reported in the literature. Based on the literature for South African populations, 1–10% of the population are PM (>2% for Black Africans and 5–10% of Caucasians) [Citation22,Citation29–30]. Therefore, not having any PM patients in our cohort falls within the expected norm. Albeit that we did not find a PM subject, one UM individual was identified that is consistent with previous reports [Citation22,Citation28].
Correlation of predicted CYP2D6 activity & reported SEs
The data were grouped into four categories to compare the predicted CYP2D6 phenotypes with participant scores for the severity of SEs. In the first category, the individual is UM and did not report any SEs. Pain was also adequately controlled. Individuals with UM status are likely to experience subtherapeutic levels of drug due to ultra-rapid metabolism. Of note, the individual was also taking indometacin; however, O-demethylation by CYP2D6 appears only to be a minor pathway [Citation31,Citation32] and therefore may not have interfered with amitriptyline metabolism in this UM patient. Taken together, low initial doses of amitriptyline, in other words, 25 mg, are not only effective but also safe and are in line with the CPIC dosing recommendations [Citation18]. Individuals with NM status (second category) did not experience adverse effects due to normal enzyme activity. The majority (64%) of those patients had an activity score of 2, which predicts normal CYP2D6 activity. Most of the NMs were receiving 25 mg of amitriptyline while three were on 50 mg and one was on 10-mg amitriptyline. These individuals were also taking a range of other medications from a minimum of 5 to a maximum of 14 including CYP2C19 inhibitors (indomethacin). There was only one NM individual whose pain was not controlled and who was not experiencing other SEs. This individual was on 25 mg of amitriptyline and was using five other medications. Since none of the other drugs interfere with CYP2D6 activity, the dose of amitriptyline could be increased in this patient for more effective pain control. The third category includes individuals with normal CYP2D6 activity but who experience SEs. Out of 17 individuals, nine have an activity score of 1, four have an activity score of 1.5 and four have an activity score of 2. Similar to category 2, the majority of these individuals were receiving 25 mg of amitriptyline; three were receiving 10 mg; two were receiving 75 mg and one individual was on 100 mg. It should also be noted that these individuals were on average taking ten other medications including CYP2D6 inhibitors (cimetidine) and CYP2C19 inhibitors (indometacin), increasing the potential for drug–drug interactions. Individuals with an activity score of 1 reported more SEs compared with those with activity scores of 1.5 and 2. This observation is consistent considering that individuals with an activity score of 1 or 1.5 are predicted to have less metabolic activity compared with individuals having an activity score of 2 [Citation18]. Among the category 3 patients, amitriptyline treatment was only effective for three individuals and the remaining patients were still experiencing pain. Therefore, amitriptyline dose should not be increased in those patients to relieve the pain, as adverse effects might worsen. Treatment with an alternative should be considered for this group of patients. The fourth category comprised two IM individuals each possessing only one decreased function allele; both subjects experienced SEs.
The data generated in this pilot investigation suggest a relationship between risk of experiencing SE(s) and decreased CYP2D6 activity. However, a number of limitations need to be considered. First, the sample size for this study was very small. As a result, there was only one UM and two IMs in our patient cohort. Second, the study cohort was genetically heterogeneous, and included individuals that were of Black African, Caucasian and Indian descent. Diverse genetic backgrounds may constitute an additional layer of complexity adding to the variability of results. Third, amitriptyline metabolism is not only dependent on CYP2D6 but also CYP2C19. Other enzymes with minor contributions may also contribute to variability especially in patients with extreme CYP2D6 and/or CYP2C19 phenotypes, in other words, UMs and PMs. Fourth, in vivo CYP2D6 and CYP2C19 metabolic capacity (i.e., actual phenotype status) should be determined to refine the relationship between genotype and phenotype and the impact of concomitant medications. The metabolizer status of patients was predicted by genotype, which may not accurately reflect the actual phenotype status of a patient. Furthermore, for some alleles activity is unknown or uncertain. Fifth, plasma concentrations of amitriptyline and its metabolites should be measured to better understand the relationship between drug level, genotype, in vivo phenotype and adverse effects to facilitate the development of an algorithm for genotype-based amitriptyline dosing for neuropathic pain. Sixth, ideally the same low-dose should be given to each patient when therapy is initiated to monitor for SEs and aid in the evaluation of whether SEs are dose dependent, impacted by the patient’s genetic makeup and/or are also influenced by other factors such as age, other health conditions or drug interactions. Finally, since the phenotypic component of the study was derived from daily questionnaires, participant subjectivity and bias are factors that may impact data interpretation. Although care was taken to ensure that the questions were phrased for easy interpretation and scoring, a certain level of subjectivity will inevitably result from self-scoring studies such as this.
Planning of large-scale definitive study
Both study objectives were met and a larger study is being planned. Based on the data generated in this pilot, a minimum cohort size of n = 180 will need to be recruited to be sufficiently powered. Our future investigation will be considerably more extensive and include, for example, phenotyping of both CYP2D6 and CYP2C19 using dextromethorphan and omeprazole as test drugs, measurement of amitriptyline plasma levels, genotyping of CYP2D6 and CYP2C19 and other pharmacogenes that are involved in the metabolism and disposition of amitriptyline and other commonly prescribed concomitant medications. It will likely be impossible, however, to completely rule out polypharmacy in a larger prospective study. When polypharmacy was included in power calculations, it was found that the use of chronic medications, in particular CYP2D6 and/or CYP2C19 inhibitors, should not affect the sample size requirements. Rather, the number of drugs taken by a patient can be used to explain some of the observed variability in pain response to amitriptyline. Furthermore, results obtained in nonpolypharmacy patients would not reflect actual ‘real-life’ patient populations and therefore, would be of limited value. In this larger study, SEs will also be captured by a self-rating questionnaire completed by the patients, which will be carefully assessed as subjective measurements may impact SE scoring.
Conclusion
In conclusion, this study has found a genotype–phenotype correlation that may potentially be clinically relevant. Subjects with predicted normal or ultra-rapid metabolizer phenotypes who are likely to have lower levels of amitriptyline, presented with fewer side effects compared to individuals with decreased or no CYP2D6 activity who are likely to have higher amitriptyline levels.
Future perspective
The findings of our pilot study support undertaking a larger scale prospective study to address the limitations discussed above. Our preliminary findings also support the use of pharmacogenetic testing in the context of amitriptyline therapy for the management of diabetic pain. Given that CYP2D6 also contributes to the metabolism of several other drugs used to treat DPN, such as duloxetine, various opioids and dextromethorphan [Citation8,Citation33], we argue that pharmacogenetic screening of CYP2D6 together with CYP2C19 will be valuable to guide drug choice and dosage and thereby improve treatment outcomes in patients with DPN. For instance, for CYP2D6 UMs an alternative drug such as gabapentin may be a better choice to avoid risk of therapeutic failure with amitriptyline. For PMs, lowering the drug dose may reduce SEs; however, if this is not sufficient, this may result in the prescription of an alternative drug.
Table 1. CYP2D6 allele frequencies in the South African diabetic peripheral neuropathy cohort sampled.
Table 2. Description of study participants based on predicted CYP2D6 phenotype and side effect profile.
An association between reported side effects to amitriptyline therapy and CYP2D6 metabolizer status was observed.
This study provides preliminary data supporting the need for pharmacogenetic testing of CYP2D6 in order to individualize amitriptyline therapy for the management of pain.
CYP2D6 gene sequencing revealed additional variation including one nonsynonymous SNP that could potentially impact enzyme function.
Ethical conduct of research
The authors state that they have obtained appropriate institutional ethics approval for this study. For investigations involving human subjects, informed consent has been obtained from the participants involved.
Supplemental Table 1
Download MS Excel (36.4 KB)Supplemental Table 2
Download MS Word (21 KB)Financial & competing interests disclosure
This study was supported by funds provided by the South African Medical Research Council, the National Research Foundation of South Africa, the National Health Laboratory Services and the Institute for Cellular and Molecular Medicine, University of Pretoria. The authors have no other relevant affiliations or financial involvement with any organization or entity on the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Additional information
Funding
References
- International Diabetes Foundation . www.idf.org .
- Boulton AJM . Management of diabetic peripheral neuropathy . Clin. Diabetes23 , 9 – 15 ( 2005 ).
- Boulton AJ , GriesFA , JervellJA . Guidelines for the diagnosis and outpatient management of diabetic peripheral neuropathy . Diabet. Med.15 , 508 – 514 ( 1998 ).
- Ulugol A , KaradagHC , TamerM , FiratZ , AslantasA , DokmeciI . Involvement of adenosine in the anti-allodynic effect of amitriptyline in streptozotocin-induced diabetic rats . Neurosci. Lett.328 , 129 – 132 ( 2002 ).
- Devi P , MadhuK , GanapathyB , SarmaG , JohnL , KulkarniC . Evaluation of efficacy and safety of gabapentin, duloxetine, and pregabalin in patients with painful diabetic peripheral neuropathy . Indian J. Pharmacol.44 , 51 – 56 ( 2012 ).
- Gore M , BrandenburgNA , HoffmanDL , TaiK-S , StaceyB . Burden of illness in painful diabetic peripheral neuropathy: the patients’ perspectives . J. Pain7 , 892 – 900 ( 2006 ).
- Attal N , CruccuG , HaanpääMet al. EFNS guidelines on pharmacological treatment of neuropathic pain . Eur. J. Neurol.13 , 1153 – 1169 ( 2006 ).
- Callaghan BC , ChengHT , StablesCL , SmithAL , FeldmanEL . Diabetic neuropathy: clinical manifestations and current treatments . Lancet Neurol.11 , 521 – 534 ( 2012 ).
- Steimer W , ZöpfK , Von AmelunxenSet al. Amitriptyline or not, that is the question: pharmacogenetic testing of CYP2D6 and CYP2C19 identifies patients with low or high risk for side effects in amitriptyline therapy . Clin. Chem.51 , 376 – 385 ( 2005 ).
- Chetty S , BaalbergenE , BhigjeeAIet al. Clinical practice guidelines for management of neuropathic pain: expert panel recommendations for South Africa . SAMJ.102 , 312 – 325 ( 2012 ).
- The Human Cytochrome P450 (CYP) Allele Nomenclature Database . www.cypalleles.ki.se/ .
- Teh LK , BertilssonL . Pharmacogenomics of CYP2D6: molecular genetics, interethnic differences and clinical importance . Drug Metab. Pharmacokinet.27 , 55 – 67 ( 2012 ).
- Zhou S-F , LiuJ-P , ChowbayB . Polymorphism of human cytochrome P450 enzymes and its clinical impact . Drug Metab. Rev.41 , 89 – 295 ( 2009 ).
- Gaedigk A , SangkuhlK , Whirl-CarrilloM , KleinT , LeederJS . Prediction of CYP2D6 phenotype from genotype across world populations . Genet. Med.19 , 69 – 76 ( 2017 ).
- Llerena A , DoradoP , Peñas-LledóEM . Pharmacogenetics of debrisoquine and its use as a marker for CYP2D6 hydroxylation capacity . 10 ( 1 ), 17 – 28 ( 2009 ).
- Llerena A , NaranjoMEG , Rodrigues-SoaresF , Penas-LledóEM , FariñasH , Tarazona-SantosE . Interethnic variability of CYP2D6 alleles and of predicted and measured metabolic phenotypes across world populations . Expert Opin. Drug Metab. Toxicol.10 ( 11 ), 1569 – 1583 ( 2014 ).
- Caudle KE , DunnenbergerHM , FreimuthRRet al. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC) . Genet. Med.19 ( 2 ), 215 – 223 ( 2017 ).
- Hicks JK , SwenJJ , ThornCFet al. Clinical Pharmacogenetics Implementation Consortium guideline for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants . Clin. Pharmacol. Ther.93 , 402 – 408 ( 2013 ).
- Hicks JK , SangkuhlK , SwenJJet al. Clinical Pharmacogenetics Implementation Consortium Guideline (CPIC®) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update . Clin. Pharmacol. Ther. doi:10.1002/cpt.597 ( 2016 ) ( Epub ahead of print ).
- Jiang Z-P , ShuY , ChenX-Pet al. The role of CYP2C19 in amitriptyline N-demethylation in Chinese subjects . Eur. J. Clin. Pharmacol.58 , 109 – 113 ( 2002 ).
- Mayfield JA , SugarmanJR . The use of the Semmes–Weinstein monofilament and other threshold tests for preventing foot ulceration and amputation in persons with diabetes . J. Fam. Pract.49 ( 11 ), S17 – S17 ( 2000 ).
- Dodgen TM , HochfeldWE , FicklHet al. Introduction of the AmpliChip CYP450 test to a South African cohort: a platform comparative prospective cohort study . BMC Med. Genet.14 , 20 ( 2013 ).
- The Human Cytochrome P450 (CYP) Allele Nomenclature Database . www.cypalleles.ki.se/cyp2d6.htm .
- Twist GP , GaedigkA , MillerNAet al. Constellation: a tool for rapid, automated phenotype assignment of a highly polymorphic pharmacogene, CYP2D6, from whole-genome sequences . NPJ Genom. Med.1 , 15007 ( 2016 ).
- Gaedigk A , SimonSD , PearceRE , BradfordLD , KennedyMJ , LeederJS . The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype . Clin. Pharmacol. Ther.83 , 234 – 242 ( 2008 ).
- Dunn OJ . Estimation of the means of dependent variables . Ann. Math. Stat.29 ( 4 ), 1095 – 1111 ( 1958 ).
- Weisstein EW . Noncentral F-Distribution ( 2002 ). http://mathworld.wolfram.com/NoncentralF-Distribution.html .
- Dodgen TM , LabuschagneCDJ , Van SchalkwykAet al. Pharmacogenetic comparison of CYP2D6 predictive and measured phenotypes in a South African cohort . Pharmacogenomics J.16 , 566 – 572 ( 2015 ).
- Gaedigk A , CoetseeC . The CYP2D6 gene locus in South African coloureds: unique allele distributions, novel alleles and gene arrangements . Eur. J. Clin. Pharmacol.64 ( 5 ), 465 – 475 ( 2008 ).
- Wright GE , NiehausDJ , DrögemöllerBI , KoenL , GaedigkA , WarnichL . Elucidation of CYP2D6 genetic diversity in a unique African population: implications for the future application of pharmacogenetics in the Xhosa population . Ann. Hum. Genet.74 ( 4 ), 340 – 350 ( 2010 ).
- Nakajima M , InoueT , ShimadaN , TokudomeS , YamamotoT , KuroiwaY . Cytochrome P450 2C9 catalyzes indomethacin O-demethylation in human liver microsomes . Drug Metab. Dispos.26 ( 3 ), 261 – 266 ( 1998 ).
- Xie G , ZhouD , ChengK-W , WongCC , RigasB . Comparative in vitro metabolism of phospho-tyrosol-indomethacin by mice, rats and humans . Biochem. Pharmacol.85 ( 8 ), 1195 – 1202 ( 2013 ).
- Ziegler D , FonsecaV . From guideline to patient: a review of recent recommendations for pharmacotherapy of painful diabetic neuropathy . J. Diabetes Complications29 ( 1 ), 146 – 156 ( 2015 ).
