Abstract
The application of pharmacogenomics could meaningfully contribute toward medicines optimization within primary care. This review identified 13 studies describing eight implementation models utilizing a multi-gene pharmacogenomic panel within a primary care or community setting. These were small feasibility studies (n <200). They demonstrated importance and feasibility of pre-test counseling, the role of the pharmacist, data integration into the electronic medical record and point-of-care clinical decision support systems (CDSS). Findings were considered alongside existing primary care prescribing practices and implementation frameworks to demonstrate how issues may be addressed by existing nationalized healthcare and primary care infrastructure. Development of point-of-care CDSS should be prioritized; establishing clinical leadership, education programs, defining practitioner roles and responsibilities and addressing commissioning issues will also be crucial.
There is robust evidence that pharmacogenomic (PGx) testing has the potential to improve patient outcomes by improving medication efficacy and preventing adverse drug reactions (ADRs) [Citation1]. Despite overcoming several barriers to implementation over the last decade, including generation of evidence supporting the clinical utility of single gene–drug interactions and the development of clinical guidelines, there is currently no consensus on how PGx should be implemented within primary care in the UK [Citation2–7].
Approximately 90% of NHS activity occurs within the primary care setting, with 300 million consultations resulting in 1.1 billion prescription items at a cost of £9.2 billion annually. With this prescribing comes the challenges of ADRs, polypharmacy and medication adherence. Nearly half of individuals over 75 years of age are prescribed five or more medications, and 50% of medications are not taken as prescribed [Citation8–11]. These constitute enormous challenges for primary care and there is considerable interest in the potential of PGx testing to address these and contribute significantly to medicines optimization [Citation12]. Exposure to medicines for which there are PGx dosing guidelines is common in primary care. 58% of all adult patients, and 89% of those over 70 years of age, are prescribed one such medicine, with 47% prescribed two or more and 7% prescribed five or more. 19–21% of all new prescriptions are for medicines where there is a corresponding PGx dosing guideline [Citation13,Citation14].
Primary care is well placed to deliver on the promise of PGx. There is already a robust infrastructure supporting medicines optimization and prescribing. Primary care practitioners (PCPs) routinely incorporate many determinants of drug response into their prescribing practice, including hepatic and renal function, co-morbidities and current prescribed medication, using clinical decision support systems (CDSS) within primary care IT systems. As such, solutions for overcoming implementation barriers, absent in other healthcare settings such as hospitals, may already exist in primary care.
Here, we review existing implementation models for PGx testing within primary care or community settings. Economic modeling indicates multigene variants are more cost-effective than single gene or variant tests [Citation15,Citation16]. Further increasing polypharmacy in primary care and need for medicine optimization reinforces the value of multigene variants. Hence, this review will identify key issues and recommendations for implementing multigene variants panel testing in primary care.
Methods
A structured literature review was performed up to 12 October 2020 to identify studies describing implementation models utilizing a multigene PGx panel within a primary care or community setting. This includes PGx decision support and non-PGx prescribing decision support in the primary care setting. The MEDLINE database was queried via the PubMed search engine using the following Boolean search terms; ((genomic) AND (primary care)) AND (implement*).
| 1. | ((((community)) OR (primary care) AND (pharmacogen*); | ||||
| 2. | ((pharmacogen*) AND (primary care)) AND (implement*); | ||||
| 3. | (pharmacogen*) AND (primary care) AND (model); | ||||
| 4. | (Primary Care) AND (Pharmacogen*); | ||||
| 5. | (Pharmacogen*) AND (community); | ||||
| 6. | (Medicines optimization) AND (implement*); | ||||
| 7. | (Medicines optimization) AND (decision support). | ||||
Articles in the English language, research article (review, clinical or observational study) and relating to humans were included. In the first instance, abstracts were reviewed to determine eligibility and, where necessary, the full manuscript was reviewed. Where there was uncertainty around eligibility, the papers were discussed between the authors.
A data collection form extracted key characteristics of the studies, as listed in . Following extraction, key themes were collated. The key themes were then mapped against two existing implementation frameworks (): IGNITE framework (Implementing GeNomics In PracTicE) and Integrating Primary and Specialist Healthcare in the UK [Citation17,Citation18].
Results
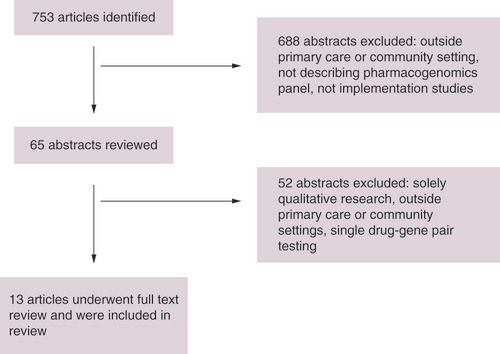
Following literature search, 753 articles were identified. As indicated in the PRISM flowchart (), 688 studies were excluded at title and abstract and 52 excluded at full text review for non-primary care setting, solely qualitative methodology or describing utilization of single-gene or drug-gene pair testing strategy [Citation19]. A total of 13 studies describing eight implementation models for PGx profiling via panel testing within primary care were identified and these are summarized in [Citation20–32]. No studies describing implementation of other genomic testing within primary care were identified. In addition two implementation frameworks describing implementation of genomic medicine and integrating non-genomic healthcare, and an implemented non-PGx Clinical Decision Support System were identified [Citation17,Citation18,Citation33].
Dressler et al. recruited three PCPs and provided education to support PGx testing within a rural primary care practice in the USA [Citation20]. Pre-emptive testing was initiated by the PCP for 49 patients who were >65 years old, had Medicare insurance, and were taking four or more prescription drugs where at least one drug had a significant drug–gene interaction. Results were returned to the PCP and pharmacist, and a summary comprising results, concurrent medications and recommendations regarding medication management was formulated by the pharmacist for use within the PCP consultation. The model was evaluated through pre- and post-testing surveys for both PCPs and patients, and impact on current medications [Citation20,Citation21].
Dunnenberger et al. implemented a multi-disciplinary clinic comprising clinical geneticist, genetic counselor and pharmacist in the USA [Citation22]. PCPs that had undergone education referred into the clinic, and pre-emptive testing offered to 76 patients, there were no exclusions. Patients had pre-test and results appointments at the clinic, each lasting an hour; the second delivered by pharmacist or clinical geneticist, and supported by a clinical summary given to the patient in addition to the electronic medical record (EMR). This model was then adjusted to offer direct access testing: following referral by the PCP, the patient was sent a testing kit comprising swab and on-line education, and the results sent to the patient and PCP and added to the EMR [Citation23]. The model was evaluated through post-test semi-structured interview of 15 PCPs [Citation23].
Schwarz et al. used a research pharmacist to identify and recruit 50 patients from one primary care practice in the USA who were on seven or more medications, or referred by their PCP [Citation25]. Consent and a buccal sample were obtained by the pharmacist, and pre-emptive testing carried out via a 15-gene panel. Pharmacogenetic data and patient-specific data were entered into a CDSS then integrated with guidelines to produce a summary and recommendations. The summary and gene card were sent to the PCP, and the gene card to the patient. Their approach was termed medication therapy management plus (MTM-Plus) and was evaluated through identification of drug–gene interactions and up-take of pharmacist recommendations by the PCP.
Haga et al. compared delivery through recruitment by a pharmacist for reactive testing after prescription of a new targeted medication with on-site support for PCPs, with recruitment by a PCP who can access support by contacting an on-call pharmacist [Citation26]. The test comprised six genes predicted to affect metabolism of 12 medications; 63 tests were performed. Results were communicated to patients at PCP discretion supported by drug-specific brochures. Evaluation was through pre- and post-test surveys of knowledge, attitudes, confidence and perceived value of PGx testing eight participating PCPs, rates of pharmacist consultation and PGx test utilization [Citation26,Citation27].
Dawes et al. identified patients with one of a list of chronic conditions, and recruited 191 participants opportunistically from PCPs in Canada [Citation28]. Testing via buccal swab and a panel, comprising 24 variants in seven genes predicted to affect metabolism of 24 medications was carried out in primary care by PCPs, and the results linked to a medication-decision support system (MDSS) and presented in a report. The MDSS was condition-linked, and PGx and biophysical data manually entered into the MDSS and combined with information from Clinical Pharmacogenetic Implementation Consortium (CPIC) guidelines to make recommendations regarding medication options and dosage. The report included related drugs and predicted phenotype. Outcomes were measured: ability to obtain and genotype samples, yield of samples, ability to link results to MDSS and utilization of MDSS.
Papastergiou et al. trained pharmacists in community pharmacies in Canada with medical geneticist support, recruiting patients on at least one medication from a known drug-gene pair opportunistically for pre-emptive testing [Citation29]. Testing via buccal swab for variants within ten genes predicted to affect metabolism of over 100 medications was carried out, and a report comprising only PGx data with recommendations based on CPIC guidelines was issued. Outcomes were rate of drug therapy problems and acceptance rates of recommendations by physicians.
Bank et al. recruited patients from primary care in The Netherlands who were issued an incident prescription from a specified list of medications from known drug-gene pairs, and had had a previous issue of any medication [Citation30]. A cohort of 200 patients out of 1114 identified as eligible were recruited by treating pharmacists, and testing via buccal swab for variants within a panel of eight genes. A report summarizing genotypes and recommending clinical action based on Dutch Working Pharmacogenomic Group (DWPG) guidelines was sent to the requesting pharmacist. The primary outcome was feasibility. Secondary outcomes were frequency of actionable genotypes (genotype on the basis of which drug prescription should be altered), number of patients with a combination of actionable genotype in combination with a drug constituting a known drug-gene pair, number of drug-related interventions and visits to the pharmacy, and drug dose. A side study quantified impact via number of PGx variants recorded in the EMR and frequency of ‘re-use’ of PGx information [Citation31].
Van der Wouden et al. utilized community pharmacists in the Netherlands who have applied PGx testing within the PREPARE (PREemptive Pharmacogenomic Testing for Preventing Adverse Drug Reactions) study of the Ubiquitous Pharmacogenomics Consortium (U-PGx). Pharmacists enrolled patients under their care, with planned initiation of one of 39 drugs with a DPWG recommendation, and conducted testing via buccal swab for variants within a panel of 12 genes. Actionable results were sent to the requesting pharmacist who then exercised discretion in whether or how to use DPWG recommendations, whether to discuss results with the treating GP and how to report the results to patients. 19 pharmacists who managed at least two actionable PGx results were recruited: mixed-methods using surveys and semi-structure interviews were utilized. Study aims were first to investigate shared decision-making, report of results to patients and time allocation, and secondly to identify remaining implementation barriers and enablers [Citation31].
All articles included were small feasibility studies describing implementation models in clinical settings, ten in North America and two in the Netherlands. Four US models comprised 49–76 patient samples and 3–15 PCPs across 1–6 primary care practice sites [Citation20–27], two Canadian models sampled 191 patients across six practice sites, and 100 patients recruited from community pharmacies respectively [Citation28,Citation29]. Two Dutch models sampled 200 patients recruited from primary care centers and recruited 19 pharmacists who applied PGx testing within their routine primary care, respectively [Citation30–32]. Outcomes primarily sought to prove feasibility of models, providing measurement of test utilization, clinical action and physician experience and these studies therefore provide useful, mostly qualitative, data to consider in implementation of PGx within primary care. summarizes study characteristics with respect to year of publication, country, setting, methodology, implementation models and outcomes.
Table 1. Summary of implementation models characteristics: recruitment strategy, patient eligibility, PGx testing strategy, PGx panel, lead practitioner, supporting practitioners, modality of results delivery, outcomes.
The literature review did not identify any existing implementation models for genomic testing in primary care. The IGNITE framework for implementation of genomic medicine interventions in clinical care identifies high priority criteria within domains, including organizational readiness for implementation, practitioner self-efficacy, knowledge and beliefs about the intervention, intervention advantage and cost and process issues [Citation17]; these priorities are mirrored by facilitators of integration of primary and specialist healthcare [Citation18]. summarizes these high priority criteria and facilitators, mapping them against themes identified from the studies describing implementation models, UK infrastructure as an exemplar of a nationalized healthcare system and potential enablers.
Table 2. Summary of results: key barriers and challenges, key enablers and solutions.
Discussion
Variation in patient populations, eligibility, sample sizes, study design, panel choice and outcome measures mean direct comparison of studies is challenging; economic evaluation was not included. North American healthcare models differ significantly from the UK in infrastructure and funding, so there may be limits of generalizability; the Dutch healthcare model is more comparable to the UK primary healthcare system.
Key influencing factors emerged from the models: pre-test counseling, PGx testing, the role of the pharmacist, recording of pharmacogenomic data, decision support, clinical outcomes and education.
Pre-test counseling
Consent was taken by a pharmacist or PCP within routine clinical care, a research or community pharmacist, or medical geneticist or genetic counselor within a genetics clinic, subsequently by sending a link to an on-line video to patients at home [Citation20,Citation22,Citation23,Citation25,Citation27–30,Citation32]. Of the latter two approaches, patients perceived neither to be of significantly different value but were more likely to undergo testing if empowered by pre-test knowledge [Citation24,Citation25].
A genetics clinic model lacked scalability; length of clinic appointment, and family and medication history taking were barriers [Citation22]. Genetic counselors have expertise in disease-based family histories which are detailed and time consuming, but have limited knowledge of prescribing [Citation22,Citation34]. PGx-relevant family history comprises only two necessary questions: ‘what medicines can a family member not take because they have caused problems in the past?’, and ‘what drug allergies do your family members have?’ [Citation22].
Time taken to consent and lack of appropriate reimbursement were significant barriers to adoption in primary care, for primary care practitioners, community pharmacists and for patients who declined testing [Citation25,Citation30,Citation32]. A condensed consent process within existing working practices is more likely to be implementable within the time-constrained primary care setting; development of a framework analogous to existing competency frameworks could be an enabler [Citation35]. PGx data is arguably more equivalent to non-genetic variables affecting prescribing than to other genetic tests and therefore demands less information than a disease-based genomic test, for example, regarding ethical issues such as implications for family members and potential for discrimination [Citation22,Citation34,Citation36–38].
Although there is currently no consensus core elements needed to reach an informed decision about PGx testing have been proposed: purpose of testing and role of genes in drug response, test risks and benefits, limitations and alternatives, future benefits and explanation that testing involves analysis of DNA [Citation39]. The lifetime nature of test results, the changing nature of guidance regarding prescribing recommendations, advice regarding DNA storage and privacy issues have been specifically suggested for inclusion [Citation40,Citation41].
Practitioner education
Continuing medical education (CME) courses on genetics and PGx are an effective way to introduce new applications and even increase personal interest, yet practitioner education was identified as a challenge with only a small number of practitioners attending educational sessions which were a requirement for participation; UK GPs favor shorter length sessions accessed on-line [Citation20,Citation42–44]. ‘Comprehensive’ community pharmacist training was regarded as essential; seminar attendance and study involvement of practitioners led to increased confidence offering, interpreting and utilizing PGx testing, but did not translate into a subsequent increase in the number of PGx tests undertaken [Citation20,Citation27,Citation29].
PCPs are more likely to access information once they see it benefiting daily practice requiring concise, relevant primary care focused resources in trusted and familiar on-line sites [Citation44–46]. Some barriers to the integration of genetics services into primary care are present in the context of PGx implemenation including lack of knowledge and guidelines [Citation47]. Point-of-care tools could raise awareness, provide education and address other barriers; guideline and protocol development has been identified as a prerequisite on which awareness and effective education can be built [Citation44,Citation47]. Provider education was identified as an on-going need and is likely to be most successful in a variety of formats including face-to-face ‘in-services’, case studies and on-line training [Citation23]. Inclusion in UK primary care curricula is currently limited [Citation48–50].
Feasibility or pilot studies can facilitate a championing model through establishing adoption, raising awareness and providing healthcare professional participants with additional knowledge [Citation20,Citation32]. Medicine Therapy Management (MTM) Plus approach involved a pharmacist with additional PGx expertise providing advice. There is broad acceptance that clinical champions are crucial to effective healthcare-related implementation, and to sustainability of clinical innovation [Citation51,Citation52]. There is also a need for specialty-specific clinical leadership to be linked to formal education programs and systemically harnessed [Citation53]. An American Society of Health-System Pharmacists (ASHP) position statement advocates pharmacist champions, pharmacists with additional training to provide leadership in development of process, guidelines and CDSS; the UK Consultant Pharmacist credentialing provides a potential mechanism [Citation54,Citation55].
Approach to PGx testing
Testing via saliva samples obtained by pharmacists or patients is feasible, with a high (99.5%) rate of genotype returns [Citation23,Citation25,Citation27–30]. PCPs varied as to whether they thought testing should be reactive (offered at the time of new prescription) or pre-emptive [Citation21]. Inability to request a test via an EMR, and a lack of established eligibility criteria were barriers to testing in primary care [Citation21]. Eligibility criteria for testing exist, and nationally-agreed testing criteria for the UK are currently under development within the National Genomic Test Directory (GTD) through an electronic test request platform [Citation56].
Of the studies where turnaround times were reported, average times of 11.7, 7, and 23 days were viewed as a challenge to implementation, although some studies found that turnaround times did not significantly affect workflow [Citation21,Citation23,Citation25,Citation27,Citation30]. Widespread pre-emptive testing could address this challenge.
PGx panels costing $102 Canadian, $300 US and $250 US were offered at no cost to the patient [Citation21,Citation25,Citation28]. Cost to the patient or medical insurer was identified as a potential barrier and reimbursement for pharmacist time was identified as imperative for implementation [Citation21,Citation25,Citation29,Citation32]. Cost and cost–effectiveness are identified as key priorities for successful implementation for PGx and genomic medicine interventions, and any intervention spanning primary and secondary care [Citation17,Citation18].
Role of the pharmacist
Arguably the most striking theme emerging from all the models was the strong support from PCPs for the involvement of pharmacists. Their varied roles included identification of eligible patients, formulation of summaries utilizing PGx data alongside other patient data, recording PGx data in the EMR, in utilization of PGx data within a full MTM consultation, either in primary care or community pharmacy settings [Citation20–32]. Testing supported via established in-house pharmacists indicated better outcomes than testing via PCP with access to an on-call pharmacist, although this model may not be deliverable at scale within the US, many PCPs in the UK already utilize pharmacist support either through community pharmacist provision, in-house or across local networks [Citation27]. 96 and 68% of PGx panel results (96 and 68%) were recorded in the community pharmacist and GP EMR respectively, although this may be because pharmacists were initiating testing [Citation32]. Furthermore, integration of PGx testing into medication reviews with pharmacist interpretation, within an appropriate pathway including GP support is feasible and community pharmacists have the confidence, capability, motivation and expertise to lead in implementation of PGx profiling into clinical practice [Citation21,Citation25,Citation26,Citation29,Citation32]. The role of the pharmacist has been recognized by professional bodies internationally, in particular in the USA where the majority of pharmacogenomics services are pharmacist led: the ASHP position statement argues that pharmacists have a fundamental responsibility to ensure PGx testing is utilized in medicines optimization, echoed in the UK with a call for pharmacist training in PGx in order to integrate into medicines optimization and medicines review processes [Citation54,Citation57].
Existing guidelines for medicine optimization recommend applying principles of PINCER (pharmacist-led Information technology intervention) to reduce medication-errors in general practice through identification of patients at risk of ADR using IT methodology with follow-up pharmacist support, feedback and educational outreach [Citation58,Citation59]. Pharmacist-led intervention in the form of medication reviews reduces adverse events in the form of hospital admissions, evidence for effectiveness of a pharmacist-only intervention in reducing medication errors is conflicting [Citation58,Citation60]. Addition of educational outreach, established working relations with practices and focus on specific examples resulted in a multi-faceted intervention more likely to be effective than single interventions [Citation58].
Recording of PGx data in the EMR
Integration of genomic information into electronic prescribing systems has been described as crucial, a pre-requisite, a ‘sine que non’ [Citation27,Citation58,Citation61]. Digitalisation of tools dramatically reduces practitioner time, with lack of or inadequate information technology (IT) infrastructure and recording of PGx data identified as barriers; ideally data would be integrated into the EMR directly, either from the laboratory or via a nation-wide EMR sharing structure [Citation21,Citation25,Citation32].
There are limitations to patient-held PGx data; only 25–35% of PGx direct-to-consumer reports are shared with physicians, and hand-held cards with PGx data and a code linked to on-line guidance were acceptable in providing clinical decision support in various healthcare settings only where EHR or IT infrastructure was lacking [Citation62,Citation63]. Barriers to use were time, difficulties integrating into routine workflow, data privacy, inability to integrate PGx with other patient data, and lack of specificity from guidelines and lack of advice regarding alternative medications [Citation64].
Decision support
PGx data must be used in conjunction with other variables affecting prescribing such as body mass index (BMI), renal and hepatic function, concurrent medications and other medical conditions [Citation65]. A point-of-care electronic CDSS or Medication Decision Support System (MDSS) integrating PGx data with other patient data and existing guidelines to make prescribing recommendations represents physician preference and is likely to serve as an educational resource; CDSS has been identified as feasible and critical to successful implementation of PGx into routine patient care [Citation26,Citation66,Citation67].
PCPs wanted decision support when receiving and discussing PGx test results; PCPs required support in report interpretation, preferring clinical summaries which include PGx data, current medications and recommendations for medication management [Citation23,Citation26,Citation32]. Guidance for utilization of PGx data in prescribing decisions exists, with concordance between CPIC and DPWG guidelines [Citation66,Citation68]. In addition to CPIC recommendations practitioners wanted situational and practical guidance either via a patient-specific summary produced by the practice pharmacy team or discussion between GPs and pharmacists [Citation23,Citation32]. Time constraints and managing patient expectations were additional challenges for PCPs [Citation23,Citation32]. A condition-based PGx MDSS in primary care integrating patient data with PGx data to give medication and dosage recommendations is feasible and accessed regularly, being utilized 236-times for 190 patients over 3 months for 11 of the 15 conditions [Citation28].
PGx CDSS have been shown to reduce hospital admissions caused by ADRs, CDSS developed outside the primary care setting have been described and utilized, but not comprehensively evaluated [Citation69]. Consideration of an existing non-PGx CDSS implemented into primary care provides insights into implementation of PGx CDSS. CDSS have previously been shown to improve practitioner prescribing performance through decreased new prescriptions for potentially harmful medication but did not affect discontinuation rate of pre-existing medications [Citation75]. The STOPP-START tool (Screening tool for older people’s potentially inappropriate prescriptions-Screening tool to alert doctors to right/appropriate treatments) is utilized within medication review and embedded within national medicines optimization guidance [Citation33,Citation59]. Algorithms run parallel to the EMR, extracting relevant patient data and recommending clinical actions.
Evidence underpinning STOPP-START implementation shows striking commonality with PGx CDSS. GPs appreciated that STOPP START allowed systematic review of the patient’s therapy, aiding prescription of required medications and withdrawal of ‘useless’ drugs and improved effectiveness when compared with a non-digitalized medication review [Citation33,Citation61]. Linkage with the EMR was regarded as absolutely necessary [Citation73]. CDSS needed to incorporate medical conditions and age, up-to-date medications, and drug-drug interactions, and should be accredited by professional organizations [Citation73]. A decisive factor influencing GPs views was relevance and validity of criteria: they should be evidence-based, up-to-date, comprise risk and benefit of prescribing decisions and be associated with a decreased risk of adverse events [Citation73]. Information was regarded as ‘useless’ if advised caution, or did not give novel information. A barrier to use was time taken to discuss changes with patients [Citation73].
Thus other factors impacting on implementation may be generalized to PGx CDSS: challenges to algorithm construction including achieving consensus, code availability and training regarding appropriate codes were critical to the STOPP-START algorithm’s success [Citation76,Citation77]. Alerts, either suggesting testing, or post-test, need to be implemented carefully to prevent alert fatigue and avoid liabilities; alert systems have variable effects on prescribing decisions [Citation69–71]. PCPs are more engaged if alerts are relevant and clinically actionable; tailoring allows time-saving and avoids alert fatigue [Citation61,Citation72–74]. STOPP-START has been implemented and embedded within national guidance despite limited evidence of economic and quality of life impact; limited evidence for clinical utility remains a barrier for implementation of PGx CDSS [Citation2,Citation33,Citation59]. Yet it has been argued that the level of evidence for PGx testing should be the same non-PGx testing when considering inclusion into guidelines and drug labelling [Citation38].
Evidence of clinical impact
Existing models provide proof of feasibility for PGx implementation in primary care, with observations regarding clinical outcomes in combination with qualitative evaluation of stakeholder’s perspectives. Further research into prescribing decisions based on PGx data, clinical utility and cost–effectiveness is required in order to support scalability.
PGx test results from 90–97% of tested individuals included actionable variants and 24–31% of patients were on a medication associated with an actionable variant (gene–drug interaction, GDI), consistent with existing data [Citation13,Citation21,Citation25,Citation27,Citation28,Citation30]. There was variability in rates at which recommendations translated into medication changes: 5% from in-house pharmacist, 63% from community pharmacists, and 90% via a MTM summary were accepted by PCPs or pharmacists [Citation25,Citation27,Citation29,Citation30]. Barriers were lack of access to PCP by pharmacists, PCP lack of understanding of applicability and non-integration into workflows [Citation29]. 97% of PGx panel results pre-recorded in the EMR were re-used at least once, with 33% used for up to four prescriptions in a 2.5 year follow-up [Citation31]. GDI seemingly had no significant effect on dosages, interventions and visits; these outcomes were secondary end-points [Citation30,Citation31].
Physicians saw value and utility in individualizing treatment, guiding decision-making, increased patient satisfaction and adherence; only three of eight providers were likely to continue to offer PGx testing with no further PGx testing offered within 6-month follow-up [Citation27].
Implementation
Nationalised commissioning, service delivery, research and educational infrastructure has the potential to facilitate implementation; for example in the UK the NHS, Genomics England, Genomics Medicine Service, GLH and the Genomic Test Directory provide strategic leadership and a robust model of care through construction of an evidence-based PGx panel, patient eligibility criteria, and delivery of direct-access testing through accredited laboratories, while HEE GEP is mandated for education and workforce planning. maps the domains and priorities identified within the two implementation frameworks against key issues identified within this review, existing infrastructure and potential enablers. The findings are consistent recent qualitative studies which identified the need for evidence for clinical utility, clear cost–effectiveness and reimbursement mechanisms, clear roles and responsibilities, clear principles for data sharing and well-developed informatics and CDSS [Citation32,Citation78].
Table 3. Frameworks for implementation of genomic and non-genomic interventions into clinical care: domains and priorities mapped against implementation issues for PGx and potential enablers.
PGx data shows greater similarity to non-genomic data used to inform prescribing decisions than to other genomic data, with implications for testing, consent and utilization. CDSS are feasible, would allow integration and alignment into existing clinical workflows and improve practitioner performance. Guidelines aiding prescribing decisions based on PGx data already exist, and, when used at point-of-care, also provide education. Challenges in developing CDSS remain, namely developing consensus, use of alerts to maximize PCP engagement, availability of PGx codes, integration of PGx data into the EMR across all care settings, and with other patient data and up-to-date guidelines to provide comprehensive prescribing decision support.
Conclusion
Review of existing implementation models provide insights into factors influencing operationalization of PGx testing into primary care. Delivery of PGx testing in primary care and community settings is feasible, with evidence for pharmacist-led delivery. A proposed delivery model within a nationalized healthcare system will need to be feasible within the resource-constrained primary care setting and be adequately commissioned. Evidence for clinical utility and cost–effectiveness are needed, and evidence for scalability is essential, facilitated by national research infrastructure with consideration for the level of supporting evidence. Awareness-raising and education programs built on CDSS, creation of clinical champions with additional expertise, pharmacy leadership with multi-disciplinary collaborative working and clear definition of roles and responsibilities, and development of clinical pathways with access to clinical expertise are all likely to support successful implementation.
Future perspective
Harnessing existing nationalized healthcare service and healthcare infrastructure could address key remaining issues in implementing PGx testing within a primary care setting. In addition development of CDSS and data integration align with strategic aims of service integration and development of secure and interoperable IT architecture. PGx data shows similarity to non-genomic data used to inform prescribing decisions, meaning existing primary care infrastructure and prescribing practices provide a foundation for integration and implementation. This similarity also has implications for development of consent and testing processes and future evidence base. The expertise and integral role of the pharmacist within the multi-disciplinary primary care team lends itself naturally to the growth of pharmacist-led PGx testing and delivery with supporting responsibilities, competencies and accreditation. CDSS are feasible, would align with current clinical practice, allow integration into clinical workflows, improve practitioner performance and provide point-of-care education through linkage with relevant guidelines. Challenges in developing CDSS remain: achieving consensus, use of alerts to maximise PCP engagement, availability of codes for PGx data and integration of PGx data into the EMR across all care settings, with other patient data and up-to-date guidelines to provide comprehensive prescribing decision support. Nevertheless implementation of PGx testing within Primary Care settings is potentially within reach and continues to provide great hope for addressing the growing challenges of polypharmacy and medicines optimization within a resource-constrained healthcare system.
PGx in primary care
Primary care practitioners (PCPs) and pharmacists within primary care already work closely to play a fundamental role in prescribing and medicines optimization.
Pharmacogenomics (PGx) has potential to contribute, however, the evidence base is lagging.
Implementation of PGx is feasible within primary care
Implementation studies demonstrate models for pre-test counseling, the role of the pharmacist and decision support including point-of-care clinical decision support systems integrated with patient information and current guidelines to give prescribing recommendations.
Existing prescribing infrastructure including non-PGx Clinical decision support system (CDSS) and the primary care multi-disciplinary approach to prescribing should be regarded as foundation for implementation of PGx testing and data into primary care.
There are additional considerations for implementation including development of pharmacist and PCP with additional expertise, definition of roles and responsibilities, defined clinical pathways, adequate reimbursement and provision of education through CDSS and educational programs.
Challenges & future directions
Implementation of PGx into primary care requires further evidence for clinical utility, cost–effectiveness and scalability.
Integration of PGx data with other patient data into the electronic medical record across all care settings, with guidelines and decision support in order to provide robust prescribing recommendations.
Existing prescribing infrastructure and implementation frameworks provides valuable insights into considerations and enablers for implementation of PGx into primary care settings.
Author contributions
J Hayward and N Qureshi conceived the article. J Hayward drafted the article with input from JH McDermott and WG Newman. JH McDermott and WG Newman offered critical input and reviewed all drafts. N Qureshi reviewed the final draft. All authors reviewed and agreed upon the final draft for submission.
Financial & competing interests disclosure
J Hayward was supported by University of Manchester and Health Education England Genomics Education Programme. The work of WG Newman is supported by the Manchester NIHR BRC (IS-BRC-1215-20007). J Hayward has a non-remunerated role with Congenica. The authors would like to thank North West Genomics Medicine Service Alliance for supporting this work. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Additional information
Funding
References
- Pirmohamed M . Personalized pharmacogenomics: predicting efficacy and adverse drug reactions. Annu. Rev. Genomics Hum. Genet.15(1), 349–370 (2014).
- Swen JJ , HuizingaTW , GelderblomHet al. Translating pharmacogenomics: challenges on the road to the clinic. PLoS Med.4(8), e209 (2007).
- Pirmohamed M , BurnsideG , ErikssonNet al. A randomized trial of genotype-guided dosing of warfarin. N. Engl. J. Med.369(24), 2294–2303 (2013).
- Turner RM , PirmohamedM. Statin-related myotoxicity: a comprehensive review of pharmacokinetic, pharmacogenomic and muscle components. J. Clin. Med.9(1), 22 (2019).
- Swen J , WiltingI , de GoedeAet al. Pharmacogenetics: from bench to byte. Clin. Pharmacol. Ther.83(5), 781–787 (2008).
- Relling MV , KleinTE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin. Pharmacol. Ther.89(3), 464–467 (2011).
- Guo C , XieX , LiJet al. Pharmacogenomics guidelines: current status and future development. Clin. Exp. Pharmacol. Physiol.46(8), 689–693 (2019).
- Alfirevic A , PirmohamedM. Genomics of adverse drug reactions. Trends Pharmacol. Sci.38(1), 100–109 (2017).
- Health Survey for England, 2016 – NHS Digital. https://digital.nhs.uk/data-and-information/publications/statistical/health-survey-for-england/health-survey-for-england-2016
- NHS England – Primary care. https://www.england.nhs.uk/five-year-forward-view/next-steps-on-the-nhs-five-year-forward-view/primary-care/
- Prescriptions Dispensed in the Community – Statistics for England, 2007–2017 – NHS Digital. https://digital.nhs.uk/data-and-information/publications/statistical/prescriptions-dispensed-in-the-community/prescriptions-dispensed-in-the-community-england---2007---2017
- The King’s Fund . Polypharmacy and medicines optimization: making it safe and sound (2013). https://www.kingsfund.org.uk/publications/polypharmacy-and-medicines-optimisation
- Kimpton JE , CareyIM , ThreapletonCJDet al. Longitudinal exposure of English primary care patients to pharmacogenomic drugs: an analysis to inform design of pre-emptive pharmacogenomic testing. Br. J. Clin. Pharmacol.85(12), 2734–2746 (2019).
- Youssef E , KirkdaleCL , WrightDJ , GuchelaarH , ThornleyT. Estimating the potential impact of implementing pre-emptive pharmacogenetic testing in primary care across the UK. Br. J. Clin. Pharmacol.87( 7), 2907–2925 (2021).
- Health economics of pharmacogenomics – Professor Dyfrig Hughes. http://www.uk-pgx-stratmed.co.uk/index.php/health-economics-of-pharmacogenomics-professor-dyfrig-hughes.html
- Plumpton CO , PirmohamedM , HughesDA. Cost–effectiveness of panel tests for multiple pharmacogenes associated with adverse drug reactions: an evaluation framework. Clin. Pharmacol. Ther.105(6), 1429–1438 (2019).
- Orlando LA , SperberNR , VoilsCet al. Developing a common framework for evaluating the implementation of genomic medicine interventions in clinical care: the IGNITE Network’s Common Measures Working Group. Genet. Med.20(6), 655–663 (2018).
- Kozlowska O , LumbA , TanGD , ReaR. Barriers and facilitators to integrating primary and specialist healthcare in the United Kingdom: a narrative literature review. Future Healthc. J.5(1), 64–80 (2018).
- PRISMA-P Group , MoherD , ShamseerLet al.Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst. Rev.4(1), 1 (2015).
- Dressler LG , BellGC , AbernathyPM , RuchK , DenslowS. Implementing pharmacogenetic testing in rural primary care practices: a pilot feasibility study. Pharmacogenomics20(6), 433–446 (2019).
- Dressler LG , BellGC , RuchKD , RetamalJD , KrugPB , PaulusRA. Implementing a personalized medicine program in a community health system. Pharmacogenomics19(17), 1345–1356 (2018).
- Dunnenberger HM , BiszewskiM , BellGCet al. Implementation of a multidisciplinary pharmacogenomics clinic in a community health system. Am. J. Health. Syst. Pharm.73(23), 1956–1966 (2016).
- Lemke AA , HuttenSelkirk CG , GlaserNSet al. Primary care physician experiences with integrated pharmacogenomic testing in a community health system. Pers. Med.14(5), 389–400 (2017).
- Lemke AA , HulickPJ , WakeDTet al. Patient perspectives following pharmacogenomics results disclosure in an integrated health system. Pharmacogenomics19(4), 321–331 (2018).
- Schwartz EJ , TurgeonJ , PatelJet al. Implementation of a standardized medication therapy management plus approach within primary care. J. Am. Board Fam. Med.30(6), 701–714 (2017).
- Haga SB , AllenLaPointe NM , ChoAet al. Pilot study of pharmacist-assisted delivery of pharmacogenetic testing in a primary care setting. Pharmacogenomics15(13), 1677–1686 (2014).
- Haga SB , MillsR , MoaddebJ , AllenLaPointe N , ChoA , GinsburgGS. Primary care providers’ use of pharmacist support for delivery of pharmacogenetic testing. Pharmacogenomics18(4), 359–367 (2017).
- Dawes M , AloiseMN , AngJSet al. Introducing pharmacogenetic testing with clinical decision support into primary care: a feasibility study. CMAJ Open4(3), E528–E534 (2016).
- Papastergiou J , ToliosP , LiW , LiJ. The Innovative Canadian Pharmacogenomic Screening Initiative in Community Pharmacy (ICANPIC) study. J. Am. Pharm. Assoc.57(5), 624–629 (2017).
- Bank PCD , SwenJJ , SchaapRD , KlootwijkDB. Baak – Pablo R, GuchelaarH-J. A pilot study of the implementation of pharmacogenomic pharmacist initiated pre-emptive testing in primary care. Eur. J. Hum. Genet.27(10), 1532–1541 (2019).
- van der Wouden CH , BankPCD , ÖzokcuK , SwenJJ , GuchelaarH-J. Pharmacist-initiated pre-emptive pharmacogenetic panel testing with clinical decision support in primary care: record of PGx results and real-world impact. Genes10(6), 416 (2019).
- van der Wouden CH , PaasmanE , TeichertM , CroneMR , GuchelaarH-J , SwenJJ. Assessing the implementation of pharmacogenomic panel-testing in primary care in the netherlands utilizing a theoretical framework. J. Clin. Med.9(3), 814 (2020).
- Hill-Taylor B , SketrisI , HaydenJ , ByrneS , O’SullivanD , ChristieR. Application of the STOPP/START criteria: a systematic review of the prevalence of potentially inappropriate prescribing in older adults, and evidence of clinical, humanistic and economic impact. J. Clin. Pharm. Ther.38(5), 360–372 (2013).
- Callard A , NewmanW , PayneK. Delivering a pharmacogenetic service: is there a role for genetic counselors?J. Genet. Couns.21(4), 527–535 (2012).
- Facilitating genomic testing: a competency framework – Genomics Education Programme. https://www.genomicseducation.hee.nhs.uk/consent-a-competency-framework/
- Fargher EA , EddyC , NewmanWet al. Patients’ and healthcare professionals’ views on pharmacogenetic testing and its future delivery in the NHS. Pharmacogenomics8(11), 1511–1519 (2007).
- Relling MV , AltmanRB , GoetzMP , EvansWE. Clinical implementation of pharmacogenomics: overcoming genetic exceptionalism. Lancet Oncol.11(6), 507–509 (2010).
- Pirmohamed M , HughesDA. Pharmacogenetic tests: the need for a level playing field. Nat. Rev. Drug Discov.12(1), 3–4 (2013).
- Mills R , VooraD , PeyserB , HagaS. Delivering pharmacogenetic testing in a primary care setting. Pharmacogenomics Pers. Med.6, 105–112 (2013).
- Weitzel KW , DuongBQ , ArwoodMJet al. A stepwise approach to implementing pharmacogenetic testing in the primary care setting. Pharmacogenomics20(15), 1103–1112 (2019).
- Robertson JA . Consent and privacy in pharmacogenetic testing. Nat. Genet.28(3), 207–209 (2001).
- Clyman JC , NazirF , TarolliS , BlackE , LombardiRQ , HigginsJJ. The impact of a genetics education program on physicians’ knowledge and genetic counseling referral patterns. Med. Teach.29(6), e143–e150 (2007).
- Houwink EJ , van TeeffelenSR , MuijtjensAMet al. Sustained effects of online genetics education: a randomized controlled trial on oncogenetics. Eur. J. Hum. Genet.22(3), 310–316 (2014).
- Evans WRH , TranterJ , RafiI , HaywardJ , QureshiN. How genomic information is accessed in clinical practice: an electronic survey of UK general practitioners. J. Community Genet.11(3), 377–386 (2020).
- Mathers J , GreenfieldS , MetcalfeA , ColeT , FlanaganS , WilsonS. Family history in primary care: understanding GPs’ resistance to clinical genetics – qualitative study. Br. J. Gen. Pract.60(574), e221–e230 (2010).
- Qureshi N . Continuous medical education approaches for clinical genetics: a postal survey of general practitioners. J. Med. Genet.39(11), 69e–669 (2002).
- Mikat-Stevens NA , LarsonIA , TariniBA. Primary-care providers’ perceived barriers to integration of genetics services: a systematic review of the literature. Genet. Med.17(3), 169–176 (2015).
- Primary Care Genetics Curriculum – primary care genetics. https://www.primarycaregenetics.org/?page_id=399&lang=en
- Clinical topic guides. https://www.rcgp.org.uk/training-exams/training/gp-curriculum-new/clinical-topic-guides.aspx
- Interim Foundation Pharmacist curriculum. https://www.rpharms.com/development/credentialing/foundation/interim-foundation-pharmacist-curriculum
- Miech EJ , RattrayNA , FlanaganME , DamschroderL , SchmidAA , DamushTM. Inside help: an integrative review of champions in healthcare-related implementation. SAGE Open Med.6, 2050312118773261 (2018).
- Martin GP , WeaverS , CurrieG , FinnR , McDonaldR. Innovation sustainability in challenging health-care contexts: embedding clinically led change in routine practice. Health Serv. Manage. Res.25(4), 190–199 (2012).
- Slade I , BurtonH. Preparing clinicians for genomic medicine: table 1. Postgrad. Med. J.92(1089), 369–371 (2016).
- Haidar C-E . Pharmacist’s Role in Clinical Pharmacogenomics. American Journal of Health-System Pharmacy72(7), 579–581 (2015).
- Consultant Pharmacist Credentialing. https://www.rpharms.com/development/credentialing/consultant/consultant-pharmacist-credentialing#how
- Education Precision Prescribing. https://test.genxys.com/content/education-materials/
- Wright D , BhattD. Targeted medicines: how pharmacists can lead a pharmacogenomics revolution. Clin. Pharm.10(6), 162–164 (2018).
- Avery AJ , RodgersS , CantrillJAet al. A pharmacist-led information technology intervention for medication errors (PINCER): a multicentre, cluster randomised, controlled trial and cost–effectiveness analysis. Lancet379(9823), 1310–1319 (2012).
- Medicines optimization: the safe and effective use of medicines to enable the best possible outcomes. https://www.nice.org.uk/guidance/ng5
- Royal S . Interventions in primary care to reduce medication related adverse events and hospital admissions: systematic review and meta-analysis. Qual. Saf. Health Care15(1), 23–31 (2006).
- Hicks JK , DunnenbergerHM , GumpperKF , HaidarCE , HoffmanJM. Integrating pharmacogenomics into electronic health records with clinical decision support. Am. J. Health. Syst. Pharm.73(23), 1967–1976 (2016).
- van der Wouden CH , CarereDA , Maitland-vander Zee AHet al. Consumer perceptions of interactions with primary care providers after direct-to-consumer personal genomic testing. Ann. Intern. Med.164(8), 513 (2016).
- Darst BF , MadlenskyL , SchorkNJ , TopolEJ , BlossCS. Characteristics of genomic test consumers who spontaneously share results with their health care provider. Health Commun.29(1), 105–108 (2014).
- Blagec K , RomagnoliKM , BoyceRD , SamwaldM. Examining perceptions of the usefulness and usability of a mobile-based system for pharmacogenomics clinical decision support: a mixed methods study. PeerJ4, e1671 (2016).
- Haga SB , MoaddebJ. Comparison of delivery strategies for pharmacogenetic testing services. Pharmacogenet Genomics24(3), 139–145 (2014).
- Relling MV , EvansWE. Pharmacogenomics in the clinic. Nature526(7573), 343–350 (2015).
- Dunnenberger HM , CrewsKR , HoffmanJMet al. Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Annu. Rev. Pharmacol. Toxicol.55(1), 89–106 (2015).
- Bank P , CaudleK , SwenJet al. Comparison of the Guidelines of the Clinical Pharmacogenetics Implementation Consortium and the Dutch Pharmacogenetics Working Group. Clin. Pharmacol. Ther.103(4), 599–618 (2018).
- Hinderer M , BoekerM , WagnerSAet al. Integrating clinical decision support systems for pharmacogenomic testing into clinical routine – a scoping review of designs of user-system interactions in recent system development. BMC Med. Inform. Decis. Mak.17(1), 81 (2017).
- Bell GC , CrewsKR , WilkinsonMRet al. Development and use of active clinical decision support for preemptive pharmacogenomics. J. Am. Med. Inform. Assoc.21(e1), e93–e99 (2014).
- Weingart SN , TothM , SandsDZ , AronsonMD , DavisRB , PhillipsRS. Physicians’ decisions to override computerized drug alerts in primary care. Arch. Intern. Med.163(21), 2625 (2003).
- Ranji SR , RennkeS , WachterRM. Computerised provider order entry combined with clinical decision support systems to improve medication safety: a narrative review. BMJ Qual. Saf.23(9), 773–780 (2014).
- Dalleur O , FeronJ-M , SpinewineA. Views of general practitioners on the use of STOPP&START in primary care: a qualitative study. Acta Clin. Belg.69(4), 251–261 (2014).
- Jeffries M , PhippsDL , HowardRL , AveryAJ , RodgersS , AshcroftDM. Understanding the implementation and adoption of a technological intervention to improve medication safety in primary care: a realist evaluation. BMC Health Serv. Res.17(1), 196 (2017).
- CMAJ . The medical office of the 21st century (MOXXI): effectiveness of computerized decision-making support in reducing inappropriate prescribing in primary care. https://www.cmaj.ca/content/169/6/549.abstract
- Huibers CJA , SalleveltBTGM , de GrootDAet al. Conversion of STOPP/START version 2 into coded algorithms for software implementation: a multidisciplinary consensus procedure. Int. J. Med. Inf.125, 110–117 (2019).
- de Groot DA , de VriesM , JolingKJet al. Specifying ICD9, ICPC and ATC codes for the STOPP/START criteria: a multidisciplinary consensus panel. Age. Ageing43(6), 773–778 (2014).
- Rafi I , CrinsonI , DawesM , RafiD , PirmohamedM , WalterFM. The implementation of pharmacogenomics into UK general practice: a qualitative study exploring barriers, challenges and opportunities. J. Community Genet.11(3), 269–277 (2020).
