Abstract
Aim: To identify novel genetic variants predisposing to elevation of Alanine aminotransferase (ALT) in rheumatoid arthritis (RA) patients after initiation of methotrexate (MTX) treatment. Patients & methods: We performed genome-wide association studies in 198 RA patients starting MTX. Outcomes were maximum level of ALT and ALT >1.5-times the upper level of normal within the first 6 months of treatment. Results:RAVER2 (rs72675408) was significantly associated with maximum level of ALT (p = 4.36 × 10-8). This variant is in linkage disequilibrium with rs72675451, which is associated with differential expression of JAK1 and RAVER2. Conclusion: We found an association between ALT elevation and genetic variants that may regulate the expression of JAK1 and RAVER2. JAK1 encodes a janus kinase involved in the pathogenesis of RA.
Low-dose methotrexate (MTX) is well-established for the treatment of rheumatoid arthritis (RA), both in monotherapy and in combination with other drugs. Ever since the introduction of MTX, the potential risk of hepatotoxicity has been of major concern, and guidelines for regular laboratory measurements for its early detection have been developed, focusing mainly on elevation of alanine aminotransferase (ALT) or aspartate aminotransferase [Citation1]. Elevation of aminotransferases is reported in more than 30% of MTX-treated RA patients and can lead to modification or withdrawal of treatment [Citation2,Citation3].
Currently, there is no way to predict MTX-induced hepatotoxicity prior to treatment start. Pharmacogenetic studies have identified potential risk factors in RA patients treated with low-dose MTX. Genetic variation in MTHFR has been associated with overall toxicity [Citation4], and we reported a tentative association with elevation of ALT [Citation5]. A tandem repeat in the enhancer region of TYMS has been studied for risk of MTX toxicity with conflicting results [Citation6,Citation7]. There is some support for a role for SLCO1B1 in MTX-related toxicity [Citation8]. Finally, a recent study reported an association between hepatotoxicity and the ADORA3 gene in the adenosine pathway, through which MTX exerts its anti-inflammatory effects [Citation9].
Furthermore, it has been speculated that MTX-induced hepatotoxicity and nonalcoholic fatty liver disease (NAFLD) might have a common pathogenic background. In genome-wide association studies (GWAS), NAFLD has been associated with PNPLA3, GCKR, SAMM50, GATAD2A, HERPUD2 and with intergenic regulatory region variants on chromosome 16 [Citation10–14]. Large GWAS in several populations have also detected genetic associations with elevation of ALT, in particular with PNPLA3 [Citation15–17].
To our knowledge, no GWAS of hepatotoxicity induced by low-dose MTX has previously been carried out in patients with RA. In this GWAS, the primary aim was to identify novel genetic risk factors associated with early hepatotoxicity after initiation of MTX as assessed by elevation of ALT within 6 months of starting treatment. A secondary outcome was association with maximum level of ALT within 6 months after treatment start. We also aimed to replicate findings from genetic studies of MTX-induced hepatotoxicity, elevation of ALT and NAFLD. Polymorphisms in MTHFR, TYMS and SLCO1B1 have been reported previously [Citation5], and these genes were therefore not included in the current study.
Materials & methods
Discovery cohort description
All patients who fulfilled the 1987 or 2010 American College of Rheumatology criteria for RA [Citation18,Citation19] and started oral or subcutaneous MTX treatment between 1 January 2005 and 30 April 2013, at the Rheumatology department at Uppsala University Hospital, Sweden, were identified from electronic health records and asked to participate. The patients were required to be at least 18 years of age and to provide written informed consent. Patient characteristics (age at onset of RA, sub-classification of RA, comorbidities and history of ALT-elevation), and details about therapy (duration and maximum dosage of MTX therapy, and time to first elevation of ALT) were retrospectively obtained from medical and laboratory records. Patients were followed from start of MTX until treatment stop, or until 30 September 2013, whichever occurred first. Data on BMI, smoking habits and alcohol consumption (measured as standard glasses per week) were collected in a telephone interview using a standardized questionnaire. ALT tests were performed according to Swedish guidelines, in other words, every 14 days during the first 3 months of MTX therapy, followed by monthly testing for 3 months, and finally every 3 months for as long as MTX therapy was maintained (https://svenskreumatologi.se/). ALT elevation was defined as ALT >1.5-times the upper limit of normal (ULN) (>44 U/l [0.75 μkat/l] in adult females, and >66 U/l [1.1 μkat/l] in adult males). The primary end point elevation of ALT >1.5 × ULN within the first 6 months of treatment was selected based on a previous study [Citation20], and patients were divided into cases and controls. In our secondary analysis, the outcome was maximum value of ALT in all patients within the first 6 months of treatment. All discovery cohort patients provided a blood sample that was kept at -70°C until DNA extraction. DNA was extracted according to standard procedures.
Statistical analyses of clinical data
Descriptive data were expressed as mean ± standard deviation, minimum (min), maximum (max) and frequency (%). For comparative analyses between cases and controls, Student’s t-test or Mann–Whitney U-test were used for continuous variables, and chi-square or Fisher’s exact test for categorical variables.
Power calculation
Statistical power was estimated using the R package GeneticsDesign (function GPC) using a prevalence of 18/198 for the disease. For a marker with an allele frequency of 0.4, there was 80% power to detect an odds ratio of approximately 7.5 at a genome-wide level (Supplementary Figure 1).
Genome-wide array data & analyses
Patients were genotyped with the Illumina Infinium OmniExpressExome 1 M Array. Genotype calls were generated using the Genome Studio software from Illumina and the Genome Reference Consortium human assembly GRCh37. PLINK v1.9 was used for genotyping quality control and data management. Principal component analysis was performed on nonimputed data in order to account for possible population stratification. Imputation was performed on the Sanger imputation server. The haplotype reference consortium panel was used as reference for the pipeline with Eagle2 (v2.0.5) prephasing and PBWT imputation. After imputation and quality control, the total number of SNPs was 7,585,873.
PLINK v1.9 was used for logistic regression analysis on a genome-wide level, and the analyses were adjusted for sex, age and the first four principal components [Citation21]. Analysis of the secondary outcome max ALT within 6 months was performed in 194 patients using linear regression implemented in PLINK v1.9. Prior to the analysis, max ALT was log2 transformed due to the right tail skewed distribution of the variable (Supplementary Figure 2). The analysis was adjusted for age, sex, log2 baseline ALT (ALT at start of MTX treatment) and the first four genetic principal components. SNP effects were modeled as additive and the statistical significance was set at the traditional genome-wide level p < 5 × 10-8 to correct for multiple testing [Citation22].
Replication cohort description
Patients diagnosed with RA [Citation18,Citation19] who started oral or subcutaneous MTX treatment at the Rheumatology Department, Uppsala University Hospital, Sweden, between 1 May 2013 and 30 September 2017, and the Rheumatology Department, Sunderby Hospital (Lulea), Sweden, between 1 January 2005 and 30 September 2017 were identified from electronic health records. Participating patients were followed from the start of MTX until treatment stop or for a minimum of 6 months. The same patient characteristics, details about the MTX therapy, and laboratory data as for the patients in the discovery cohort were collected. All patients provided a blood or saliva sample (2 ml Oragene® OG-500 collection kit, DNA Genotek, Canada). All samples were kept at -70°C until DNA extraction. DNA was extracted according to standard procedures.
Replication genotyping & meta-analysis
Genotyping was performed using the TaqMan SNP Genotyping Assay kit for RAVER2 rs72675408 (C__99351193_10) on the Applied Biosystems 7500 Fast Real-Time PCR System (Thermo Fisher Scientific, MA, USA) according to standard procedures. Statistical analysis of the outcome max ALT within 6 months was performed using linear regression implemented in PLINK 1.9. Prior to the analysis, max ALT was log2 transformed due to the right tail skewed distribution of the variable (Supplementary Figure 2). The analysis was adjusted for age, sex, log2 baseline ALT and the first four genetic principal components. The SNP effect was modeled as additive. Meta-analysis was performed using a fixed model in the metafor R-package. The cut-off for a statistically significant association was set to p < 0.05.
Functional variants analysis
LDlink suite of applications (https://ldlink.nci.nih.gov/) was used to interrogate linkage disequilibrium (LD) between the top GWAS SNPs and other SNPs (r2 >0.6). Functional annotations were obtained by intersecting the obtained SNPs with chromatin state models based on imputed data in adult liver (EID: E066) from the Roadmap Epigenome Project [Citation23]. Data on transcription factor binding were obtained from chromatin immunoprecipitation sequencing (ChIP-seq) experiments in the Encyclopedia of DNA Elements (ENCODE) project [Citation24]. Association of SNPs with gene activity in different tissues was calculated using the expression Quantitative Trait Loci (eQTL) calculator tool from the Genotype-Tissue Expression (GTEx) project [Citation25]. The HiCap method was used to assess physical 3D contacts between enhancers and promoters [Citation26].
Candidate gene analysis
In the imputed dataset, we examined 14 candidate SNPs that previously have been implicated in studies on MTX-induced hepatotoxicity, elevation of ALT and NAFLD. The following variants were investigated: the ADORA3 haplotype rs2298191T/rs1544223A/rs3393A [Citation9], rs4808199 in GATAD2A [Citation11], rs1260326 and rs780094 in GCKR [Citation11,Citation12], rs10272006 in SP4 [Citation13], rs343064 near HERPUD2 [Citation14], rs738409 and rs2896019 in PNPLA3 [Citation11–16], rs738491 and rs2143571 in SAMM50 [Citation11], and rs6499186 and rs698718, both intergenic on chromosome 16 [Citation13]. Bonferroni correction was used to adjust for multiple testing for 12 independent tests, and the cut-off for statistical significance was set to p < 0.0042 (0.05/12).
Results
Discovery cohort characteristics
A total of 213 RA patients starting MTX treatment were included in the discovery cohort, of whom 15 were later excluded (three due to known non-MTX related elevation of ALT, three did not provide DNA samples and nine did not provide any ALT test within the first 6 months after initiation of MTX treatment). Characteristics of the remaining 198 patients are shown in .
Table 1. Characteristics of patients initiating methotrexate treatment included in the discovery and replication cohorts.
Eighteen patients (9%) were defined as cases due to at least one elevation of ALT >1.5 × ULN within the first 6 months, while the remaining 180 were defined as controls (Supplementary Table 1). Ethnic origin did not differ significantly between cases and controls (p = 0.46, Supplementary Table 1). Both parents of 83% of the cases and 87% of the controls were born in Sweden. One outlier was detected with principal component analysis (Supplementary Figure 3). This person was defined as a control (ALT ≤1.5 × ULN). Sensitivity analysis excluding the outlier did not change the results, and the outlier was therefore not removed from the study.
Four patients, who were reported to have normal ALT throughout the first 6 months, were excluded from analysis of maximum ALT because their exact ALT values were not recorded. All patients were treated with MTX 7.5–25 mg once weekly and were supplemented with folate. When comparing cases with controls, the proportion of individuals with a previous history of elevation of ALT before MTX treatment was significantly higher among cases compared with controls (50 vs 12.5%, p < 0.001). The mean duration of MTX treatment was significantly shorter in cases than controls (3.0 years vs 4.4 years, p = 0.02), and the mean maximum dose of MTX was significantly lower (15.1 vs 17.3 mg, p = 0.045).
Genome-wide association analysis, primary outcome
No genome-wide statistically significant association was found when comparing patients with ALT >1.5 × ULN within 6 months with controls (, Supplementary Figure 4 & Supplementary Table 2). Tentative associations (p < 2 × 10-5) were in or close to CDC123, RAVER2, and HNF1B.
Table 2. Top 6 genome-wide results for alanine aminotransferase above 1.5-times the upper limit of normal within 6 months.
Genome-wide association analysis, secondary outcome
The maximum level of ALT within 6 months was significantly associated with four SNPs in high LD in or close to RAVER2 at a genome-wide level (, & Supplementary Table 3). The RAVER2 intron SNP rs72675408 had a beta-value of 0.83 per increase of one minor allele (). This means that the geometric mean of ALT increased 1.78-times per minor allele of rs72675408. In other words, an ALT of 1.00 would be predicted to increase to 1.78 in patients with one variant allele, and to (1.78)2 = 3.17 in patients with two variant alleles. Three SNPs located upstream of RAVER2 also passed correction for multiple testing: rs3920617, rs55889764 and rs17384589 ().
Alanine aminotransferase (ALT) is log2 transformed and adjusted by log (baseline ALT), age, sex and genetic principal components 1-4. The line denotes the significance level p < 5 × 10-8.
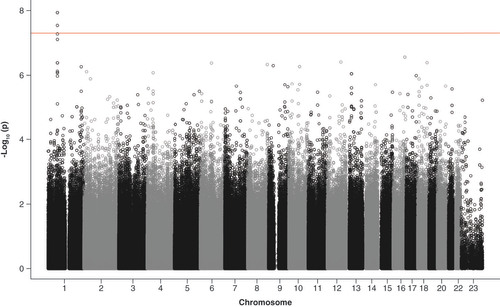
Table 3. Top 6 genome-wide results for maximum alanine aminotransferase within 6 months.
Replication & meta-analysis
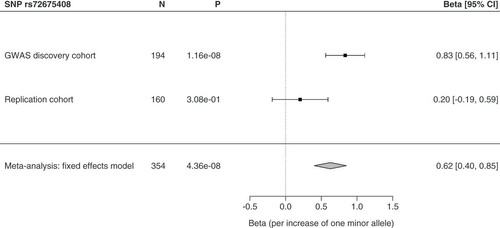
Patient characteristics of the replication cohort (n = 160) are shown in . We sought replication for the top RAVER2 hit rs72675408. The variant showed a tendency in the same direction in the replication cohort, but did not reach statistical significance (). Meta-analysis of the discovery and replication cohorts (n = 354) gave a significant result (beta per increase of one minor allele of rs72675408 = 0.62, 95% CI = 0.40–0.85, p = 4.36 × 10-8) ().
Meta-analysis was performed using random and fixed effects models in the metafor R-package. SNP effects were modeled as additive. Estimated odds ratios with 95% CI, p-value and numbers (N) included are shown. The cut-off for a statistically significant association was p < 0.05.
Functional analysis
The four significantly associated SNPs are in high LD with one another, and with several others (r2 >0.6) in the JAK1 and RAVER2 locus (Supplementary Figure 5). Most of these SNPs are located in intronic and noncoding parts, whereas three of them, rs3737139, rs2230586 and rs11585932, are synonymous coding mutations in JAK1. Data from the GTEx project [Citation25] show that rs72675451 and rs17392542 are eQTLs for JAK1 and RAVER2 in the thyroid, and rs72675451 has a similar trend for RAVER2 in the liver (Supplementary Table 4A & B). In addition, rs56345619 and rs17392542 are eQTLs for RAVER2 in diverse tissues. Based on data from the Roadmap Epigenome Project [Citation23], the intron variants rs72675451 in JAK1 and rs12402976 in RAVER2 are located in enhancers active in the liver (Supplementary Table 4A & B). Data generated using the HiCap method [Citation26] indicate that rs72675451 makes physical 3D contacts with the promoters of JAK1 and RAVER2 in the liver [Citation27], suggesting that this polymorphic enhancer may regulate both genes (). In summary, functional analyses indicate that an enhancer harboring rs72675451 might regulate both JAK1 and RAVER2.
The enhancer harboring rs72675451 interacts with JAK1 and RAVER2 promoters in human liver tissue (pink lines) [Citation27]. The insert shows transcription factors binding in ChIP-seq experiments from the Encyclopedia of DNA Elements project [Citation24]. The color intensity is proportional to signal strength (abbreviations at: http://tinyurl.com/watv2v7).
![Figure 3. The genomic landscape for the SNPs rs72675408, rs72675451 and rs12402976.The enhancer harboring rs72675451 interacts with JAK1 and RAVER2 promoters in human liver tissue (pink lines) [Citation27]. The insert shows transcription factors binding in ChIP-seq experiments from the Encyclopedia of DNA Elements project [Citation24]. The color intensity is proportional to signal strength (abbreviations at: http://tinyurl.com/watv2v7).](/cms/asset/3949e156-8296-4e7c-8319-1e13f5e0647c/ipgs_a_12349029_f0003.jpg)
Candidate gene analyses
When cases with ALT >1.5 × ULN were compared with controls, no statistically significant association was revealed (Supplementary Table 5). Maximum level of ALT within 6 months was nominally associated with the GATAD2A intron variant rs4808199, but the result did not pass correction for multiple testing ().
Table 4. Candidate SNP results for maximum alanine aminotransferase within 6 months.
Discussion
Hepatotoxicity is a potentially serious adverse effect of low-dose MTX, and for this reason, guidelines recommend regular ALT testing. However, most of the tests are normal; in a previous study, it has been reported that only 7% of ALT tests were pathologic, in other words, above ULN [Citation3]. Methods to predict elevation of ALT during low-dose MTX treatment are currently lacking. Identifying patients at risk before treatment start could offer a possibility to personalize treatment, and focus ALT testing on susceptible patients.
In this GWAS, the RAVER2 gene was tentatively associated with ALT >1.5 × ULN, and significantly associated with maximum level of ALT within 6 months of initiation of MTX. RAVER2 encodes a heterogeneous nuclear ribonucleoprotein that interacts with the polypyrimidine tract binding protein (PTB) by participating in its nuclear functions or modulating its activity [Citation28]. PTB is involved in all steps of mRNA metabolism and acts as a repressor of alternatively spliced exons [Citation29]. RAVER2 is ubiquitously expressed in the body, but its expression in the liver is low [Citation25,Citation30]. An in vitro study in human embryonic stem cells showed that folic acid deprivation induced by MTX led to differential expression of several genes encoding RNA-binding proteins, among them RAVER2 [Citation31]. A candidate gene study has further reported an association between the RAVER2 variant rs2780814 and ulcerative colitis [Citation32].
When investigating SNPs in high LD with the four top hits, we identified one (rs72675451) located in an enhancer [Citation23] that makes physical 3D contacts with the promoters of JAK1 and RAVER2 in the liver [Citation27]. In addition, it is an eQTL for both genes in the thyroid, with a similar trend for RAVER2 in the liver [Citation25]. JAK1 encodes one of the janus kinases (JAKs) that play an important role in intracellular signaling pathways involved in the pathogenesis of RA [Citation33]. When used in RA therapy, MTX partly acts as an anti-inflammatory agent suppressing the JAK signal transduction pathway of several interleukins [Citation34]. Furthermore, specific JAK inhibitors currently used in the treatment of RA (tofacitinib, baricitinib and upadacitinib) may cause adverse liver effects [Citation35–37]. The above findings suggest that rs72675451 is a functional variant that may regulate JAK1 and RAVER2, and thereby influence MTX treatment response.
The possible relevance of polymorphisms in genes associated with NAFLD and/or elevation of ALT has to our knowledge not previously been investigated in patients with RA treated with MTX. NAFLD is part of the metabolic syndrome, which is prevalent among patients with RA and associated with increased disease activity [Citation38]. This could support a common pathogenesis or that MTX treatment could unmask a pre-existing NAFLD [Citation38,Citation39]. We did not find any significant association between genes previously associated with NAFLD and/or elevation of ALT. The study participants with an average BMI of 26.5 (range 17–41), and a prevalence of diabetes of 7.2% were, however, not a typical high-risk group for NAFLD.
The main limitation of this GWAS is the small study size. Our study was powered to detect common SNPs with relatively high odds ratios, and uncommon SNPs conferring a small risk could therefore go undetected. However, gene variants associated with adverse effects generally have larger effect sizes than variants predisposing to disease, and consequently significant results may be obtained with smaller cohorts [Citation40]. Sequencing to detect multiple rare risk variants could be a future project, but is outside the scope of the current study. Another limitation is that functional findings should be confirmed experimentally. Strengths are that the study is representative for the contemporary Swedish RA population based on sex, age and treatment, and that the detected association was supported by meta-analysis of two cohorts.
Conclusion
We detected an association between early elevation of ALT during low-dose MTX treatment and SNPs that may regulate the expression of the JAK1 and RAVER2 genes. If these findings are replicated, they could be used in a polygenic risk score for prediction of patients at risk of hepatotoxicity when starting low-dose MTX therapy.
Low-dose methotrexate (MTX) is an established treatment for rheumatoid arthritis.
Liver toxicity is a well-known complication of MTX treatment.
Patients with rheumatoid arthritis starting MTX were identified from electronic health records.
Their liver enzyme alanine aminotransferase (ALT) levels were followed for 6 months.
We performed genome-wide association studies aiming to identify novel genetic risk factors associated with ALT elevation.
Variants in and upstream of RAVER2 were significantly associated with maximum ALT within 6 months of starting MTX.
There is evidence that these variants may regulate the expression of the genes JAK1 and RAVER2.
If replicated, the findings could be used in a polygenic risk score for MTX-induced liver toxicity.
Author contributions
P Hallberg, M Wadelius, J Karlsson Sundbaum and E Baecklund contributed to study design. J Karlsson Sundbaum, E Baecklund, M Wallenberg contributed to case, control and data collection. J Karlsson Sundbaum contributed to adjudication of cases. H Kohnke assisted in genotyping. Data analysis was performed by N Eriksson, J Karlsson Sundbaum. M Cavalli and C Wadelius contributed to functional analysis. J Karlsson Sundbaum, P Hallberg, M Wadelius, N Eriksson, E Baecklund, M Cavalli, C Wadelius contributed to data interpretation. J Karlsson Sundbaum, P Hallberg, M Wadelius, N Eriksson, M Cavalli, C Wadelius, E Baecklund assisted in manuscript drafting. M Wadelius, P Hallberg, N Eriksson, J Karlsson Sundbaum, E Baecklund, M Cavalli, C Wadelius contributed to revising manuscript content. All authors approved the final version of the manuscript.
Ethical conduct of research
Research was carried out in accordance with the latest update of the Declaration of Helsinki. The study was approved by the regional ethics committee (2010/231, Uppsala, Sweden), and written informed consent was obtained from all participants.
Supplemental Figure 1
Download MS Word (745.1 KB)Acknowledgments
The authors are grateful to all physicians, research nurses and supporting staff, who assisted in recruiting patients and controls or administering phenotype databases. The authors particularly thank staff at the clinics of Rheumatology at Uppsala University Hospital and Sunderby Hospital, and RN Ulrica Ramqvist, RN Charlotta Haglund and assistant S Collin, and assistant E Prado at Uppsala University, Sweden.Genotyping of the discovery cohort was performed by the single nucleotide polymorphism and sequencing (SNP&SEQ) Technology Platform at Uppsala University (www.genotyping.se). Computations were done on resources provided by the Swedish National Infrastructure for Computing through the Uppsala Multidisciplinary Centre for Advanced Computational Science (UPPMAX). Genotyping of the replication cohort was performed at Clinical Pharmacology, Uppsala University Hospital.
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.futuremedicine.com/doi/suppl/10.2217/pgs-2021-0064
Financial & competing interests disclosure
This work was supported by the Agnes and Mac Rudberg’s, Brunnberg’s, Selander’s and Thuréus’ Foundations at Uppsala University; Swedish Rheumatism Association; Swedish Diabetes foundation (DIA2017-269); Swedish Research Council (Medicine 521-2011-2440, 521-2014-3370 and 2018-03307); Swedish Heart-Lung Foundation (20120557, 20140291 and 20170711); and Clinical Research Support (Avtal om Läkarutbildning och Forskning, ALF) at Uppsala University. The SNP&SEQ Technology Platform is part of the National Genomics Infrastructure which is supported by the Swedish Research Council for Infrastructures and Science for Life Laboratory, Sweden. The SNP&SEQ Technology Platform is also supported by the Knut and Alice Wallenberg Foundation. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Additional information
Funding
References
- Singh JA , SaagKG , BridgesSLJret al. 2015 American College of Rheumatology Guideline for the Treatment of Rheumatoid Arthritis. Arthritis Rheumatol.68(1), 1–26 (2016).
- Visser K , VanDer Heijde DM. Risk and management of liver toxicity during methotrexate treatment in rheumatoid and psoriatic arthritis: a systematic review of the literature. Clin. Exp. Rheumatol.27(6), 1017–1025 (2009).
- Karlsson Sundbaum J , ErikssonN , HallbergP , LehtoN , WadeliusM , BaecklundE. Methotrexate treatment in rheumatoid arthritis and elevated liver enzymes: a long-term follow-up of predictors, surveillance, and outcome in clinical practice. Int. J. Rheum. Dis.22(7), 1226–1232 (2019).
- Song GG , BaeS-C , LeeYH. Association of the MTHFR C677T and A1298C polymorphisms with methotrexate toxicity in rheumatoid arthritis: a meta-analysis. Clin. Rheumatol.33(12), 1715–1724 (2014).
- Karlsson Sundbaum J , BaecklundE , ErikssonN , HallbergP , KohnkeH , WadeliusM. MTHFR, TYMS and SLCO1B1 polymorphisms and adverse liver effects of methotrexate in rheumatoid arthritis. Pharmacogenomics21(5), 337–346 (2020).
- Lima A , SeabraV , BernardesM , AzevedoR , SousaH , MedeirosR. Role of key TYMS polymorphisms on methotrexate therapeutic outcome in portuguese rheumatoid arthritis patients. PLoS ONE9(10), e108165–e108165 (2014).
- Jekic B , LukovicL , BunjevackiVet al. Association of the TYMS 3G/3G genotype with poor response and GGH 354GG genotype with the bone marrow toxicity of the methotrexate in RA patients. Eur. J. Clin. Pharm.69(3), 377–383 (2013).
- Lima A , BernardesM , AzevedoRet al. SLC19A1, SLC46A1 and SLCO1B1 polymorphisms as predictors of methotrexate-related toxicity in Portuguese rheumatoid arthritis patients. Toxicol. Sci.142(1), 196–209 (2014).
- Grk M , MilicV , DolzanVet al. Analysis of association of ADORA2A and ADORA3 polymorphisms genotypes/haplotypes with efficacy and toxicity of methotrexate in patients with Rheumatoid arthritis. Pharmacogenomics J.20(6), 784–791 (2020).
- Eslam M , ValentiL , RomeoS. Genetics and epigenetics of NAFLD and NASH: clinical impact. J. Hepatol.68(2), 268–279 (2018).
- Kawaguchi T , ShimaT , MizunoMet al. Risk estimation model for nonalcoholic fatty liver disease in the Japanese using multiple genetic markers. PLoS ONE13(1), e0185490–e0185490 (2018).
- Di Costanzo A , BelardinilliF , BailettiDet al. Evaluation of polygenic determinants of non-alcoholic fatty liver disease (NAFLD) By a candidate genes resequencing strategy. Sci. Rep.8(1), 3702–3702 (2018).
- Namjou B , LingrenT , HuangYet al. GWAS and enrichment analyses of non-alcoholic fatty liver disease identify new trait-associated genes and pathways across eMERGE Network. BMC Medicine17(1), 135 (2019).
- Chalasani N , GuoX , LoombaRet al. Genome-wide association study identifies variants associated with histologic features of nonalcoholic fatty liver disease. Gastroenterology139(5), 1567–1576 (2010).
- Chambers JC , ZhangW , SehmiJet al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat. Genet.43(11), 1131–1138 (2011).
- Kanai M , AkiyamaM , TakahashiAet al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat. Genet.50(3), 390–400 (2018).
- Speliotes EK , Yerges-ArmstrongLM , WuJet al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genetics7(3), e1001324 (2011).
- Arnett FC , EdworthySM , BlochDAet al. The American rheumatism association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum.31(3), 315–324 (1988).
- Aletaha D , NeogiT , SilmanAJet al. 2010 Rheumatoid arthritis classification criteria: An American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum.62(9), 2569–2581 (2010).
- Schmajuk G , MiaoY , YazdanyJ , BoscardinWJ , DaikhDI , SteinmanMA. Identification of risk factors for elevated transaminases in methotrexate users through an electronic health record. Arthritis Care Res. (Hoboken)66(8), 1159–1166 (2014).
- Chang CC , ChowCC , TellierLC , VattikutiS , PurcellSM , LeeJJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience4, 7 (2015).
- Sham PC , PurcellSM. Statistical power and significance testing in large-scale genetic studies. Nat. Rev. Genet.15(5), 335–346 (2014).
- Kundaje A , MeulemanW , ErnstJet al. Integrative analysis of 111 reference human epigenomes. Nature518(7539), 317–330 (2015).
- Consortium EP . An integrated encyclopedia of DNA elements in the human genome. Nature489(7414), 57–74 (2012).
- Lonsdale J , ThomasJ , SalvatoreMet al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet.45(6), 580–585 (2013).
- Sahlén P , AbdullayevI , RamsköldDet al. Genome-wide mapping of promoter-anchored interactions with close to single-enhancer resolution. Genome Biol.16(1), 156 (2015).
- Cavalli M , DiamantiK , PanGet al. A multi-omics approach to liver diseases: integration of single nuclei transcriptomics with proteomics and HiCap bulk data in human liver. OMICS24(4), 180–194 (2020).
- Henneberg B , SwiniarskiS , SabineB , IllenbergerS. A conserved peptide motif in Raver2 mediates its interaction with the polypyrimidine tract-binding protein. Exp. Cell Res.316(6), 966–979 (2010).
- Bartoletti-Stella A , GaspariniL , GiacominiCet al. Messenger RNA processing is altered in autosomal dominant leukodystrophy. Hum. Mol. Genet.26(19), 3868–3868 (2017).
- Uhlén M , FagerbergL , HallströmBMet al. Tissue-based map of the human proteome. Science347(6220), 1260419 (2015).
- Liu W , WangK , LvXet al. Up-regulation of RNA binding proteins contributes to folate deficiency-induced neural crest cells dysfunction. Int. J. Biol. Sci.16(1), 85–98 (2020).
- Bouzid D , FouratiH , AmouriAet al. Association of the RAVER2 gene with increased susceptibility for ulcerative colitis. Hum. Immunol.73(7), 732–735 (2012).
- Choy EHS , Miceli-RichardC , González-GayMAet al. The effect of JAK1/JAK2 inhibition in rheumatoid arthritis: efficacy and safety of baricitinib. Clin. Exp. Rheumatol.37(4), 694–704 (2019).
- Gremese E , AliverniniS , TolussoB , ZeidlerMP , FerraccioliG. JAK inhibition by methotrexate (and csDMARDs) may explain clinical efficacy as monotherapy and combination therapy. J. Leukoc. Biol.106(5), 1063–1068 (2019).
- Mueller RB , HaslerC , PoppFet al. Effectiveness, tolerability, and safety of tofacitinib in rheumatoid arthritis: a retrospective analysis of real-world data from the St. Gallen and Aarau Cohorts. J. Clin. Med.8(10), 1548 (2019).
- Wu Z-P , ZhangP , BaiJ-Z , LiangY , HeJ-S , WangJ-C. Efficacy and safety of baricitinib for active rheumatoid arthritis in patients with an inadequate response to conventional synthetic or biological disease-modifying anti-rheumatic drugs: a meta-analysis of randomized controlled trials. Exp. Ther. Med.16(3), 2449–2459 (2018).
- Serhal L , EdwardsCJ. Upadacitinib for the treatment of rheumatoid arthritis. Expert Rev. Clin. Immunol.15(1), 13–25 (2019).
- Da Cunha VR , BrenolCV , BrenolJCTet al. Metabolic syndrome prevalence is increased in rheumatoid arthritis patients and is associated with disease activity. Scand. J. Rheumatol.41(3), 186–191 (2012).
- Langman G , HallPDLM , ToddG. Role of non-alcoholic steatohepatitis in methotrexate-induced liver injury. J. Gastroenterol. Hepatol.16(12), 1395–1401 (2001).
- Maranville JC , CoxNJ. Pharmacogenomic variants have larger effect sizes than genetic variants associated with other dichotomous complex traits. Pharmacogenomics J.16(4), 388–392 (2016).
