Abstract
Pharmacogenomics is emerging as an important component both in facilitating new drug development and in improving the utility of existing chemotherapeutic agents. Both candidate gene and genome-wide approaches have been used to identify genetic markers associated with chemotherapeutic response and/or toxicity. New molecular targeted agents have been designed based on a sophisticated understanding of the molecular alterations defining cancers. Over the next decade, the translation of these findings into clinical practice, as well as functional studies of genetic variants, is likely to take center stage. More comprehensive evaluation of the human genome, including the examination of rare SNPs, copy number variations, tandem repeats and epigenetic effects, will further improve our understanding of the relationship between genetics and drug response.
The paradigm of individualized drug therapy based on a person‘s unique genetic make-up is especially desirable in the field of oncology, where the therapeutic index is often narrow and the consequences of drug toxicity can be life threatening. Generally, anticancer agents are administered at the maximally tolerated dose, as defined for a large population, with an expectation that approximately a third of patients will have unacceptable toxicity. Polymorphisms that affect drug clearance often lead to recognizable clinical events, such as myelosuppression or neurotoxicity. Thus, if clinicians could better predict which individuals are at the greatest risk of suffering chemotherapy-related toxicities, then the overall care of cancer patients could be greatly impacted with patient-specific dose modifications, optimization of treatment choice (when several equivalent therapies exist), or avoidance of a therapy when toxicity risks outweigh potential benefits. More important is the ability to predict those patients at greatest risk for nonresponse to specific treatment, as modifying their therapeutic regimen could be highly beneficial. This commentary will focus on the methods used to identify genetic variants that have been important in chemotherapy over the past decade and expected changes over the next decade.
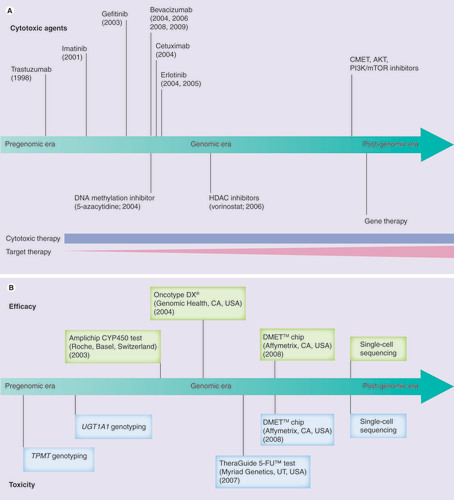
(A) Time of approval of various cancer therapies based on US FDA approval time. (B) Timeline to show the establishment of pharmacogenetic prognostic tools.
DMET: Drug-metabolizing enzymes and transporters; HDAC: Histone deacetylase.
Past
Improving cancer patient care through our understanding of the pharmacogenomics of response and toxicity of existing chemotherapy is critical; however, many existing agents are not effective and new agents are needed. Over the past decade, a number of molecularly targeted agents have been discovered based on a more sophisticated understanding of genetic and molecular processes within cancers. Examples of agents developed include trastuzumab (Herceptin®, Genentech, CA, USA), a Her-2 inhibitor, designed to target the amplified Her-2 receptors on the breast cancer cell surface Citation[1]; imatinib (Gleevec®, Novartis, Basel, Switzerland), targeting abnormal Bcr–Abl fusion protein in chronic myeloid leukemia Citation[2]; and a series of EGF receptor-tyrosine kinase inhibitors Citation[3] and anti-VEGF agents Citation[4]. Furthermore, increased knowledge in epigenetics has led to the development of novel histone deacetylase inhibitors and DNA methylation inhibitors Citation[5]. illustrates the US FDA approval time for various chemotherapeutic agents over the past decade. Although cytotoxic agents will continue to play a significant role in cancer treatment over the next decade, targeted agents will continue to be developed and are likely to become central players in clinical cancer management.
To see where approaches in the pharmacogenomics of anticancer agents are headed, it is instructive to take a step back and see how approaches emerged from the pre- to post-genomic era. In the early 2000s and for the previous decades, the approach to studing genetic variants important in response to chemotherapeutic agents was primarily via a candidate gene or pathway centric approach, a method that makes assumptions about which genes are most important in the drug‘s activity. Observations (e.g., severe toxicity) were made in patients following treatment with oncology drugs, and case–control studies were then designed to evaluate genetic variations contributing to the observed effect. Follow-up functional studies were often performed in cultured cells or animal models. Most of these studies involved a genetic variant within a drug-metabolizing gene that had a large effect on the degree or rate at which a drug was converted to its metabolites. Examples include: genetic variations in TPMT that account for more than 90% of cases with low or intermediate TPMT enzyme activity and lead to an increased risk for severe myelosuppression after 6-mercaptopurine treatment Citation[6]; UGT1A1*28, which is associated with a decrease in UGT1A1 expression and an increased risk of severe neutropenia when medium- or high-dose irinotecan is administrated Citation[7]; and lack of response to tamoxifen in CYP2D6 poor metabolizers Citation[8]. With the success of these single gene pharmacogenetic discoveries, additional prognostic tools to help guide chemotherapy based on an individual‘s somatic or germline genetic profile have been developed .
Present
Current approaches used to identify genetic variants contributing to response or toxicity associated with chemotherapy have been driven, to a greater extent, by technology that now allows us to evaluate the entire genome. This is very useful because such studies are unbiased, take into account pharmacodynamic and pharmacokinetic genes, and consider that drug response is multigenic with many genes contributing smaller effects. In contrast to the candidate gene approach, which is hypothesis driven, genome-wide approaches are considered to be hypothesis generating. However, the genome-wide approach can result in false discoveries due to multiple testing, and requires large sample sizes for reasonable power to detect associations. In oncology, replication studies are particularly challenging because the study requires individuals with the same cancer, treated in the same manner in a field where the standard of care tends to change as new therapies are tested. For these reasons and the challenges of discovery studies in humans, some have turned to cell lines. These cell-based models have shown that chemotherapeutic-induced cytotoxicity is heritable Citation[9,10] and amenable to genetic dissection Citation[9,10]. The utility of these cell-based models will certainly be better understood over the next decade.
Since the declared completion of the human genome sequence Citation[11] and the comprehensive genomic characterization of human cancers Citation[12], genome-wide association studies have gained popularity. Numerous studies employed gene-expression arrays to identify signatures that classify subtypes of various cancers Citation[13–15] and subgroups with differential sensitivity to chemotherapeutic agents Citation[16,17]. In addition to expression signatures, genetic signatures, in the form of SNPs Citation[18,19], copy number variations Citation[20,21] and mutations Citation[14], are coming to the forefront as a means to identify individuals that are likely to respond to chemotherapy. Although scientists have enjoyed a rapid advance in technology, translating laboratory findings into the clinic has been painfully slow. This is owing to the difficulty in conducting large prospective pharmacogenomic and/or pharmacoeconomic trials to demonstrate the scientific value and cost–effectiveness of pharmacogenomic prediction models. This is particularly true in the field of oncology.
Future
What will the next decade bring us in terms of genetic information? Surprisingly, the Human Genome Project demonstrated that only 2% of the human genome is actually genes Citation[11]. Therefore, researchers have begun to move beyond the scope of genes and into the regulatory sequence and the study of noncoding sequences. Intergenic, intronic and other noncoding regions may harbor regulators of gene expression Citation[22]. Meanwhile, experimental tools have included microsomes, hepatocytes and HapMap lymphoblastoid cell lines rich in genetic information Citation[23]. Over the next decade, researchers are likely to make the transition from large genome-wide association studies to a combination of genome-wide association studies and candidate gene studies, as well as validation of true positives. There will be more genetic information, in the form of rare SNPs from the 1000 Genomes Project, copy number variations and epigenetics, that will allow a better view of contributions to phenotypic variation, but which will require clever data analysis. Certainly, over the next decade we will have a much better understanding of how genetics plays a role in differences in response/toxicity to anticancer agents in individuals from different geographic locations.
While the challenges both for oncologist/hematologist and society are significant, increasing knowledge regarding the human genome and cancer genome will be concomitant with the development of comprehensive, cost-efficient and high-throughput technologies. There will certainly be an increase in powerful tools in biomedical and clinical informatics for analyzing and storing vast amounts of data, which will enable continued growth in the field of personalized therapeutics for cancer patients. For pharmacogenomics to be successful in the field of oncology, there needs to be alternative therapies with different mechanisms available for patients whose genetics show that they are at risk for severe toxicity or nonresponse with standard chemotherapy. We expect to see more ‘designer drugs‘ that are tailored to individual needs by the year 2020, along with prognostic prediction tools that can be used to guide cancer therapy.
Financial & competing interests disclosure
The authors are supported through the Pharmacogenetics of Anticancer Agents Research Group by the NIH/NIGMS grant U01GM61393, University of Chicago Breast Cancer SPORE grant P50 CA125183, and Career Development Award, University of Chicago Cancer Center Support Grant (#P30 CA14599), NCI RO1 CA136765 and R21 CA139278. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Additional information
Funding
Bibliography
- Weiner L : An overview of monoclonal antibody therapy of cancer.Semin. Oncol.26(4 Suppl. 12) , 41–50 (1999).
- Druker BJ : Translation of the Philadelphia chromosome into therapy for CML.Blood112(13) , 4808–4817 (2008).
- Ritter C , ArteagaC: The epidermal growth factor receptor-tyrosine kinase: a promising therapeutic target in solid tumors.Semin. Oncol.30(1 Suppl. 1) , 3–11 (2003).
- Jain RK , DudaDG, ClarkJW, LoefflerJS: Lessons from Phase III clinical trials on anti-VEGF therapy for cancer.Nat. Clin. Prac. Oncol.3(1) , 24–40 (2006).
- Lane AA , ChabnerBA: Histone deacetylase inhibitors in cancer therapy.J. Clin. Oncol.27(32) , 5459–5468 (2009).
- Lennard L , LilleymanJ, LoonJV, WeinshilboumR: Genetic variation in response to 6-mercaptopurine for childhood acute lymphoblastic leukaemia.Lancet336(8709) , 225–229 (1990).
- Hoskins J , GoldbergR, QuP, IbrahimJ, McleodH: UGT1A1*28 genotype and irinotecan-induced neutropenia: dose matters.J. Natl Cancer Inst.99(17) , 1290–1295 (2007).
- Goetz MP , RaeJM, SumanVJ et al.: Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes.J. Clin. Oncol.23(36) , 9312–9318 (2005).
- Dolan ME , NewboldKG, NagasubramanianR et al.: Heritability and linkage analysis of sensitivity to cisplatin-induced cytotoxicity.Cancer Res.64(12) , 4353–4356 (2004).
- Watters JW , KrajaA, Meucci Ma, Province MA, Mcleod HL: Genome-wide discovery of loci influencing chemotherapy cytotoxicity. Proc. Natl Acad. Sci. USA101(32) , 11809–11814 (2004).
- McPherson JD , MarraM, HillierL et al.: A physical map of the human genome.Nature409(6822) , 934–941 (2001).
- Cancer Genome Atlas Research Network: Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature455(7216) , 1061–1068 (2008).
- Morris S , CareyL: Gene expression profiling in breast cancer.Curr. Opin. Oncol.19(6) , 547–551 (2007).
- Thomas RK , WeirB, MeyersonM: Genomic approaches to lung cancer.Clin. Cancer Res.12(14) , S4384–S4391 (2006).
- Holleman A , Den Boer Ml, De Menezes RX et al.: The expression of 70 apoptosis genes in relation to lineage, genetic subtype, cellular drug resistance, and outcome in childhood acute lymphoblastic leukemia. Blood107(2) , 769–776 (2006).
- Holleman A , CheokM, Den Boer M et al.: Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N. Engl. J. Med.351(6) , 533–542 (2004).
- Potti A , DressmanHK, BildA et al.: Genomic signatures to guide the use of chemotherapeutics.Nat. Med.12(11) , 1294–1300 (2006).
- Huang RS , DuanS, ShuklaSJ et al.: Identification of genetic variants contributing to cisplatin-induced cytotoxicity using a genome-wide approach.Am. J. Hum. Genet.81(3) , 427–437 (2007).
- Huang Y -T, Heist RS, Chirieac LR et al.: Genome-wide analysis of survival in early-stage non-small-cell lung cancer. J. Clin. Oncol.27(16) , 2660–2667 (2009).
- Yang JJ , BhojwaniD, YangW et al.: Genome-wide copy number profiling reveals molecular evolution from diagnosis to relapse in childhood acute lymphoblastic leukemia.Blood112(10) , 4178–4183 (2008).
- Zhang Y , MartensJWM, YuJX et al.: Copy number alterations that predict metastatic capability of human breast cancer.Cancer Res.69(9) , 3795–3801 (2009).
- Duan S , HuangR, ZhangW et al.: Genetic architecture of transcript-level variation in humans.Am. J. Hum. Genet.82(5) , 1101–1113 (2008).
- International Hapmap Consortium; Frazer K, Ballinger D, Cox DR et al.: A second generation human haplotype map of over 3.1 million SNPs. Nature449(7164) , 851–861 (2007).