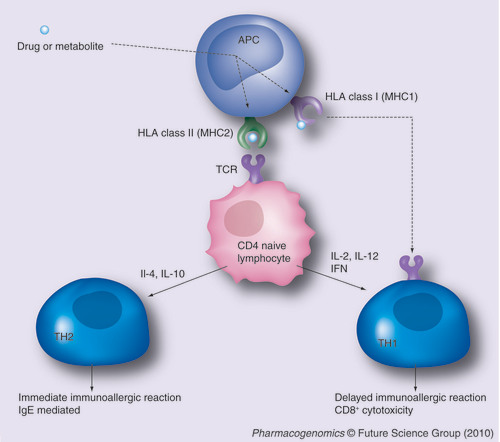
The drug or a drug metabolite is processed by an APC and presented by a class I or class II HLA protein. The HLA–drug (hapten) complex occurs only in the presence of a specific HLA allele. The presentation of the hapten to the naive CD4 lymphocyte via its TCR triggers the immunologic reaction. Depending on the cytokine immunologic environment, the activation is oriented either via a TH1 reaction (in the presence of TH1 ILs such as IL-2, IL-12 or IFN) or TH2 reaction (in the presence of TH2 interleukins such as IL-4 or IL10). Alternatively, the drug or the hapten processed by an HLA class I protein can directly stimulate cytotoxic CD8 lymphocytes.
APC: Antigen-presenting cell; IFN: Interferon; IL: Interleukin; TCR: T-cell receptor.
Adverse drug reactions (ADRs) represent a major health problem, accounting for 6–7% of hospitalization cases and approximately 100,000 deaths a year in the USA Citation[1]. They also represent the primary cause of drug withdrawal from the market Citation[2]. Most ADRs are considered type A, that is, predictable, dose dependent and related to the pharmacological action of the drugs. However, 5–20% of ADRs are unpredictable (type B), mainly related to immunoallergic reactions. From a clinical point of view, immunoallergic ADRs (IADRs) can be divided into two categories: immediate allergic events such as urticaria, anaphylaxis, collapsus and angioedema and delayed allergic events such as Stevens–Johnson syndrome (SJS), toxic epidermal necrolysis (TEN), drug rash with eosinophilia and systemic symptoms, or isolated organ injury. The former occur during the first hours after drug administration and are mainly mediated by drug-specific IgE antibodies. The latter occur several days or weeks after drug initiation and are primarily mediated by T-lymphocyte cytotoxicity.
Immunoallergic ADR symptoms are nonspecific, therefore the causal relationship with a drug is frequently difficult to establish, even when taking into account the delay between drug prescription and the event, the improvement after drug withdrawal, and recurrence after re-exposure (positive rechallenge).
The most prevalent hypothesis used to explain drug immunoallergic reactions is the hapten/prohapten theory: according to this hypothesis, the drug (or a drug metabolite) is processed by an antigen-presenting cell (APC) and further expressed at the cell membrane in the groove of a class I (MHC1) or a class II HLA (MHC2) molecule. The HLA–drug (hapten) complex is presented to a naive lymphocyte via its T-cell receptor, which will initiate different types of immunological responses, depending on the type of HLA molecule expressed by the APC and the cytokine environment . Two other hypotheses, involving either a direct interaction of the drug with the T-cell receptor and the drug Citation[3] and the cellular release of danger biological signals Citation[4], have been proposed. All three are not mutually exclusive. However, many factors that trigger the drug-related immunoallergic reaction are still missing. As a consequence, until recently there were only a few unspecific biomarkers such as eosinophilia to signal the immunoallergic nature of an ADR. Skin tests, specific IgE and in vitro lymphocyte proliferation tests can be used after the event, but their specificity or sensitivity remain disappointing Citation[5].
On the other hand, it has been known for a long time that drugs can induce immunological diseases Citation[6], such as systemic lupus erythematosus or vasculitis, which occur in susceptible patients expressing specific HLA epitopes. This indicates a genetic susceptibility to developing IADRs, restricted to HLA class I (A, B and C) or class II alleles. In addition, more and more IADRs have been identified as being associated with HLA loci since the 1970s.
However, the real ‘drug–HLA story‘ started at the beginning of the century with abacavir: in 2002, two independent groups observed that abacavir hypersensitivity syndrome was restricted to carriers of the HLA-B*5701 allele with the highest odds ratio ever met (>100) Citation[7,8]. Following this discovery, GlaxoSmithKline (London, UK) conducted the largest international pharmacogenetic randomized clinical trial ever performed to date, which was released in 2008 Citation[9]. They demonstrated that screening for HLA-B*5701 before introducing abacavir and the exclusion of the patients carrying this allele resulted in the disappearance of the hypersensitivity syndrome related to this drug, which usually occurs in 5% of the patients treated with abacavir during the first weeks of treatment. This pharmacogenetic test is now routinely used in many different countries before introducing abacavir Citation[10–12].
During the same period, screening of HLA alleles in patients hospitalized for life-threatening cutaneous ADRs (SJS and TEN), allowed for the identification of additional HLA alleles as potent risk factors for IADR. Expressing the HLA-B*1502 allele exposes patients of Asian origin to a 2504- and 36-fold higher risk of SJS to carbamazepine Citation[13] and phenytoin Citation[14], respectively . Since that time, many different HLA loci have been demonstrated to be highly predictive of specific type B ADRs related to specific drugs . HLA alleles have high negative predictive values , but low positive values to predict ADRs, indicating that these genetic biomarkers are necessary but not sufficient for the triggering of such immunoallergic events. According to the hapten theory, it is believed that the hapten complex, which triggers the immunoallergic reaction, only occurs in the presence of a specific HLA allele. Therefore, prospective HLA screening should prevent some patients from receiving a certain drug, if they express a specific risk allele, in order to avoid a potentially life threatening IADR.
The technological screening facilities mainly explain the recent and sudden identification of different HLA alleles as potent risk factors of IADRs: simultaneous screening for a large number of HLA epitopes has been performed for a long time in organ transplantation and also allowed for the early identification of HLA-restricted drug-related diseases Citation[6]. Recent genomic technologies offered the opportunity to simultaneously screen, in so-called genome-wide association studies (GWAS), most of the thousands of alleles Citation[15] of the extended human MHC Citation[16]. Furthermore, owing to the extraordinarily high risks conferred by several HLA alleles (odds ratio ranging from 10 to 2500), GWAS can be performed with less than 100 subjects and still allow for the discovery of positive pharmacogenomic hits Citation[17]. Using this agnostic genome-wide strategy, Daly et al. recently identified HLA-B*5701 as a potent risk factor for drug-induced liver injury (DILI) owing to flucloxacillin from the Drug Induced Liver Injury and Gene (DILIGEN) study, which included 51 cases and 289 controls Citation[18]. In a retrospective study performed in 74 cases and 130 controls, AstraZeneca (London, UK) was able to identify HLA-DRB1*0701 as the major genetic risk factor associated with ximelagatran DILI, which led to its withdrawal soon after its introduction on the market. This underlines the crucial need for systematic DNA banking during clinical drug development. DNA biobanks collected during drug development or in the post-marketing phase allows one, if an IARD is identified, to rapidly perform a retrospective GWAS and identify a predictive genomic biomarker. The systematic use of the latter will certainly limit the indication of the drug, but will on the other hand help to avoid its withdrawal from the market.
An increasingly more affordable genome-wide screening approach, combined with the high risk predicted by some HLA alleles, allows us to hope that in the near future, more and more severe IADRs might be prevented by pharmacogenomics testing Citation[19]. However, looking forward to that day, several issues remain to be addressed. They will be discussed here.
In order to be used in routine medical practice, the positive or negative predictive values of the HLA biomarker must reach 100%. Some of the HLA alleles listed in fulfill this condition. Even so, the cost–effectiveness of systematic screening before drug prescription must be positive. For a frequent IADR, as observed with abacavir, this condition has been verified Citation[20], but for rare events, such as allopurinol-induced SJS, it remains to be established. In addition, one should also keep in mind that a HLA risk allele may be ethnically restricted: HLA-B*1502 is highly predictive of carbamazepine-induced SJS in different Asian populations. However, in a European survey of SJS/TEN related to carbamazepine, only four out of 12 patients were carriers of the HLA-B*1502; interestingly these four patients had an Asian ancestry Citation[21]. HLA-restricted immunological response is also specific to a certain type of adverse event for a specific drug: HLA-B*1502 is a strong predictor of carbamazepine-related SJS/TEN but not of carbamazepine isolated skin rashes Citation[22], which are more frequently observed in carriers of the HLA-A*3101Citation[22]. Conversely, the same HLA allele can be predictive of different IADRs of different drugs such as HLA-B*5701, which is a strong risk factor of abacavir HSS, as well as flucloxacillin DILI.
However, despite these limits underlying the need for further research into the mechanisms of IADRs, we now have strong evidence that some IADRs are linked to specific HLA alleles and can be prevented by prospective pharmacogenomics testing. The pharmacogenomics of HLA is a recent field that has been rapidly translated into the clinic and is already improving the way we prescribe drugs. It will probably become more and more important over the next few years.
Table 1. Pharmacogenomic risk factors for adverse drug reactions.
Financial & competing interests disclosure
Laurent Becquemont has received consulting fees from Sanofi-Aventis, Pfizer, Servier and lecture fees from Genzyme, GlaxoSmithKline, Bristol–Myers Squibb, Wyeth and Merck Sharp and Dohme. His wife works for Sanofi-Aventis. The author has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript
Additional information
Funding
Bibliography
- Lazarou J , PomeranzBH, CoreyPN: Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies.JAMA279(15) , 1200–1205 (1998).
- Wysowski D , SwartzL: Adverse drug event surveillance and drug withdrawals in the United States, 1969–2002.Arch. Intern. Med.165 , 1363–1369 (2005).
- Pichler WJ : Delayed drug hypersensitivity reactions.Ann. Intern. Med.139(8) , 683–693 (2003).
- Matzinger P : Tolerance, danger, and the extended family.Annu. Rev. Immunol.12 , 991–1045 (1994).
- Torres MJ , MayorgaC, BlancaM: Nonimmediate allergic reactions induced by drugs: pathogenesis and diagnostic tests.J. Investig. Allergol. Clin. Immunol.19(2) , 80–90 (2009).
- Dedeoglu F : Drug-induced autoimmunity.Curr. Opin. Rheumatol.21(5) , 547–551 (2009).
- Hetherington S , HughesA, MostellerM et al.: Genetic variations in HLA-B region and hypersensitivity reactions to abacavir.Lancet359 , 1121–1122 (2002).
- Mallal S , NolanD, WittC et al.: Association between presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir.Lancet359(9308) , 727–732 (2002).
- Mallal S , PhillipsE, CarosiG et al.: HLA-B*5701 screening for hypersensitivity to abacavir.N. Engl. J. Med.358 , 568–579 (2008).
- Higgs J , GambhirN, RamsdenSC, PoultonK, NewmanWG: Pharmacogenetic testing in the United Kingdom genetics and immunogenetics laboratories.Genet. Test Mol. Biomarkers14(1) , 121–125 (2009).
- Watson ME , PatelLG, HaB, WannamakerP, CuffeR, ShaeferM: A study of HIV provider attitudes toward HLA-B 5701 testing in the United States.AIDS Patient Care STDS23(11) , 957–963 (2009).
- Young B , SquiresK, PatelP et al.: First large, multicenter, open-label study utilizing HLA-B*5701 screening for abacavir hypersensitivity in North America.AIDS22(13) , 1673–1675 (2008).
- Chung W , HungS, HongH et al.: Medical genetics: a marker for Stevens–Johnson syndrome.Nature428 , 486 (2004).
- Locharernkul C , LoplumlertJ, LimotaiC et al.: Carbamazepine and phenytoin induced Stevens–Johnson syndrome is associated with HLA-B*1502 allele in Thai population.Epilepsia49(12) , 2087–2091 (2008).
- Holdsworth R , HurleyCK, MarshSG et al.: The HLA dictionary 2008: a summary of HLA-A, -B, -C, -DRB1/3/4/5, and -DQB1 alleles and their association with serologically defined HLA-A, -B, -C, -DR, and -DQ antigens.Tissue Antigens73(2) , 95–170 (2009).
- Horton R , WilmingL, RandV et al.: Gene map of the extended major histocompatibility complex.Nat. Rev. Genet.5 , 889–899 (2004).
- Nelson M , Bacanu S-A, Mosteller M et al.: Genome-wide approaches to identify pharmacogenetic contributions to adverse drug reactions. Pharmacogenomics J.9(1) , 23–33 (2009).
- Daly A , DonaldsonP, BhatnagarP et al.: HLA-B*5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin.Nat. Genet.41 , 816–819 (2009).
- Becquemont L : Pharmacogenomics of adverse drug reactions: practical applications and perspectives.Pharmacogenomics10(6) , 961–969 (2009).
- Hughes DA , VilarFJ, WardCC, AlfirevicA, ParkBK, PirmohamedM: Cost–effectiveness analysis of HLA-B*5701 genotyping in preventing abacavir hypersensitivity.Pharmacogenetics14(6) , 335–342 (2004).
- Lonjou C , ThomasL, BorotN et al.: A marker for Stevens–Johnson syndrome: ethnicity matters.Pharmacogenomics J.6(4) , 265–268 (2006).
- Hung SI , ChungWH, JeeSH et al.: Genetic susceptibility to carbamazepine-induced cutaneous adverse drug reactions.Pharmacogenet. Genomics16(4) , 297–306 (2006).
- Hung S , ChungW, LiouL et al.: HLA-B*5801 allele as a genetic marker for severe cutaneous adverse reactions caused by allopurinol.Proc. Natl Acad. Sci. USA102(11) , 4134–4139 (2005).
- Donaldson P , BhatnagarP, GrahamJ et al.: Flucloxacillin-induced liver injury: the extended MHC 57.1 haplotype as a major risk factor.Hepatology48(Suppl. 4) , A396–A397 (2008).
- Martin A , NolanD, JamesI et al.: Predisposition to nevirapine hypersensitivity associated with HLA-DRB1*0101 and abrogated by low CD4 T-cell counts.AIDS19 , 97–99 (2005).
- Littera R , CarcassiC, MasalaA et al.: HLA-dependent hypersensitivity to nevirapine in Sardinian HIV patients.AIDS20 , 1624–1626 (2006).
- Kindmark A , JawaidA, HarbronC et al.: Genome-wide pharmacogenetic investigation of a hepatic adverse event without clinical signs of immunopathology suggests an underlying immune pathogenesis.Pharmacogenomics J.8 , 186–195 (2008).
- Gueant-Rodriguez RM , GueantJL, ViolaM, TramoyD, GaetaF, RomanoA: Association of tumor necrosis factor-α-308G>A polymorphism with ige-mediated allergy to betalactams in an Italian population.Pharmacogenomics J.8(2) , 162–168 (2008).
- Romano A , De Santis A, Romito A et al.: Delayed hypersensitivity to aminopenicillins is related to major histocompatibility complex genes. Ann. Allergy Asthma Immunol.80(5) , 433–437 (1998).
- O‘Donohue J , OienKA, DonaldsonP et al.: Co-amoxiclav jaundice: clinical and histological features and HLA class II association.Gut47(5) , 717–720 (2000).
- Dettling M , CascorbiI, Opgen-RheinC, SchaubR: Clozapine-induced agranulocytosis in schizophrenic Caucasians: confirming clues for associations with human leukocyte class I and II antigens.Pharmacogenomics J.7(5) , 325–332 (2007).
- Lonjou C , BorotN, SekulaP et al.: A European study of HLA-B in Stevens–Johnson syndrome and toxic epidermal necrolysis related to five high-risk drugs.Pharmacogenet. Genomics18(2) , 99–107 (2008).
- Kim SH , ChoiJH, LeeKW et al.: The human leucocyte antigen-DRB1*1302-DQB1*0609-DPB1*0201 haplotype may be a strong genetic marker for aspirin-induced urticaria.Clin. Exp. Allergy35(3) , 339–344 (2005).