Mayo Clinic Launches Whole-Genome Breast Cancer Study
Project to bring together oncologists, surgeons, radiologists, and genomics and cancer researchers to uncover the clues to effective individualized therapies and drug discovery in breast cancer.
†Biopsy utilized for tumor sequencing and generation of patient-derived xenografts.
MBI: Molecular breast imaging.
Figure courtesy of Matthew P Goetz.
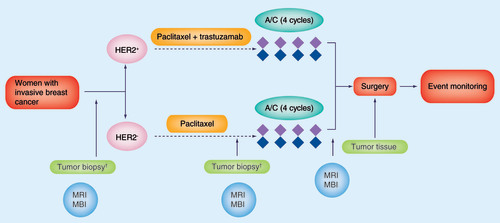
The Mayo Clinic (MN, USA)-led BEAUTY study will use blood samples, breast tumor samples and imaging, with both MRI and molecular breast imaging (MBI), to assess response in over 200 breast cancer patients undergoing a standard neoadjuvant chemotherapy regimen. It is hoped that the results will help physicians tailor chemotherapy to breast cancer patients based on their individual genomes and the genomes of their tumors.
Speaking to Pharmacogenomics about the aim of the BEAUTY project, Matthew Goetz, Mayo oncologist and study co-leader, explains: “The primary aim of this study is to identify novel somatic changes within genes and gene pathways that are potentially ‘druggable‘ in women with breast cancer undergoing a standard neoadjuvant chemotherapy regimen of paclitaxel [with or without trastuzumab], followed by A/C chemotherapy.”
An additional aim of the study is to use breast cancer tissue obtained prior to chemotherapy to develop tumor xenograft cell lines for mechanistic and functional studies to determine whether mutations identified are associated with the malignant phenotype and response to associated drugs, which target the gene and/or pathways. It is hoped that these xenograft models could also be used for future drug development.
“An additional novel aspect is the use of MBI to assess the association between changes in 99mTc-sestamibi and pathologic response following neoadjuvant chemotherapy”, highlights Goetz, who will be leading the project with Judy Boughey, a Mayo breast surgeon.
The sequencing in the study will be conducted using blood (or germline) and tumor samples, before and after chemotherapy. In addition, tumor tissue from the breast biopsy will be used to create mouse avatars, which will allow the researchers to immortalize the tumor cells. As Goetz explains: “This will allow an in vivo evaluation of potential agents matched to drug targets identified from the genomic sequencing.”
MBI with 99mTc-sestamibi will be performed prior to chemotherapy, once during chemotherapy and after completion of chemotherapy to assess how MBI can predict response to chemotherapy.
Asked how the project may have an impact on the conduct of breast cancer research and the treatment of women with breast cancer, Goetz speculates: “This study builds upon the current shift in the use of the neoadjuvant approach to evaluate new agents for breast cancer. The US FDA has endorsed the neoadjuvant setting for the preliminary approval of new drugs, using pathological complete response as the end point. This will potentially accelerate drug development, decrease cost and require fewer patients.”
Another goal of the BEAUTY project is to develop new drugs in women predicted to have poor response to standard chemotherapy: “The end result is individualized therapy – such that future treatments can be tailored to individual patients who not only exhibit high-risk disease, but who are predicted to poorly respond to standard chemotherapy.”
Going on to highlight how this study marks one of many ongoing efforts at the Mayo Clinic to personalize medicine, Goetz explains: “At Mayo we strongly believe that pharmacogenomics will continue to be the wedge that drives individualized medicine into practice, allowing us to select the right drug for the right patient at the right time, increasing both safety and effectiveness. Merging pharmacogenomics data with the electronic medical record and creating the rules engines that allow providers to take advantage of the new knowledge without having to alter their patient care flow is essential.”
Asked how the Mayo Clinic intends to drive this individualization of medicine into practice, the researcher explains: “In oncology increasingly we will utilize sequence-dependent information to guide drug selection in real time. This process may initially be slow given the smaller and smaller numbers of patients for a given genetic alteration. The challenge will be to develop novel ways to study new drugs in smaller numbers of patients.”
In a concluding statement about the present study, Goetz states: “We believe that BEAUTY will be a paradigm shift in oncology given the ability to sequence the tumor, identify genetic alterations and test the functional implication in immortalized tumor cells prior to moving to the next step. This is likely to be the way other solid tumors will be evaluated in the future.”
More information on the Mayo Clinic Center for Individualized Medicine can be found at http://mayoresearch.mayo.edu/mayo/research/center-for-individualized-medicine.
– Written by Natalie Harrison
Source: Mayo Clinic News: http://newsblog.mayoclinic.org/2012/04/05/mayo-clinic-launches-whole-genome-breast-cancer-study
Illumina launches MyGenome® iPad® app
The launch of an iPhone® application that gives users access to their own fully sequenced genome has been long awaited; at the first Consumer Genetics Show in Boston (MA, USA) in 2009, whole-genome sequencing company Illumina‘s CEO Jay Flatley showed rough concepts for an Illumina iPhone application. This month saw the launch of the company‘s MyGenome iPad application, marking another step towards the marriage of genomics and wireless health.
At the 2009 event, Flatley already had visions of genetic data being mobile-connected: “We think an iPhone-type device is where this data will end up living, but clearly we can‘t fit an entire sequence on the iPhone of today.”
While the application, which is available for download on the App StoreSM for US$0.99, does not yet include the personal genomic data for its customers, the concept is still the same. As stated in the press release, the app helps users “explore a real human genome and view reports about important genetic variations through a simple, intuitive and educational interface for genetic data exploration and learning.”
“Illumina‘s vision of a future where healthcare is made more precise through the use of genetic information, together with our position as a sequencing technology leader, puts us in an ideal position to stimulate interest in a mainstream tool for genomic exploration,” Flatley stated in the app announcement.
“The MyGenome® app is an exciting educational tool that enables consumers to learn how much we already understand about variation in the human genome, served up in a graphically accessible format. This first version of the app provides a glimpse of what we think could become a clinical tool for use by physicians with their patients to improve understanding and communication of genetic data” details Flatley.
Illumina, the leading developer, manufacturer and marketer of life science tools and integrated systems for the analysis of genetic variation and function, have future visions for the MyGenome app, which include allowing users to download their own data securely and delivering genetic data to the ordering physician. It is the hoped that physicians will be able to access a consumer‘s genomic information from the app, allowing them to visualize the genome and interrogate it for issues such as pharmacogenomic drug response, before providing direct access to the consumer.
– Written by Natalie Harrison
Source: Illumina press release: http://investor.illumina.com/phoenix.zhtml?c=121127&p=irol-newsArticle&ID=1686310
New Partnership between Horizon and the European Molecular Genetics Quality Network to Standardize Cancer Diagnostic Testing across Europe
The research tool provider Horizon Diagnostics, of Horizon Discovery Limited (Cambridge, UK), has recently announced plans for a new partnership with the European Molecular Genetics Quality Network (Manchester, UK). The partnership seeks to reduce difficulties in sourcing diagnostic testing reference materials for diagnostic external quality assessment providers. Horizon Diagnostics will now be supplying reference standard human cell lines, which contain frequencies of mutations that guide cancer therapy prescription. The cell lines will be distributed globally to molecular diagnostic laboratories participating in the European Molecular Genetics Quality Network proficiency testing schemes. Paul Morrill, Commercial Director of Horizon Discovery, elucidates the potential benefits for patients: “Quality assurance schemes such as those organized by European Molecular Genetics Quality Network are essential to ensure that patients receive the correct treatment regimen based on their tumor mutation status … this is an invaluable component of patient care.”
– Written by Jenaid Rees
Source: Horizon press release: www.horizondiscovery.com/news/article/609
New genes linked to autism
Three papers recently published in Nature reveal mutations in at least three genes that can be linked to autism spectrum disorders (ASDs) through work by the Autism Sequencing Consortium (ASC). The papers suggest that de novo mutations are a common risk factor for the development of ASDs.
Three proteins, SCN2A, CHD8 and KATNAL2 stood out as having mutations that are highly linked to a risk of developing autism. However, mutations were also discovered in more than 200 other proteins that may be involved in some way towards the development of ASDs.
Researchers from multiple institutions took part in the study, including Yale University School of Medicine (CT, USA), Harvard Medical School (MA, USA), Mount Sinai School of Medicine (NY, USA) and University of Washington School of Medicine (WA, USA).
The three studies all used whole-exome sequencing, and analysis of only the protein-coding exons of the genome, to determine mutations in genes that were found in individuals with ASDs. The combined studies looked at the genomes from 580 families with a child with an ASD, but with no family history of autism. This enabled researchers to look for de novo mutations, ones that are not found in either parent but have newly occurred in their children.
Neale et al. demonstrated that the rate of de novo mutations found throughout the genome was slightly higher in affected individuals than in their unaffected families. While many of these mutations will not affect the risk of autism, a study of the protein–protein interactions of the gene products found that proteins that had mutations were more likely to interact with each other.
The authors suggest that these results “support polygenic models in which spontaneous coding mutations in any of a large number of genes increases risk by five- to 20-fold.”
Joseph Buxbaum, from Mount Sinai School of Medicine and lead author of the study, explains that “We now have a good sense of the large number of genes involved in autism and have discovered about 10% of them.”
Mark Daly, a coauthor from Harvard Medical School, said: “These genes hold key insights into the true biological causes of autism – insights we have been unable to gain through other lines of research.”
The study by O‘Roak et al. found that mutations were much more likely to be passed from the father, with paternal mutations four-times as prevalent as maternal ones. This confirms previous theories that austism risk has a paternal bias.
The third study compared results of individuals with ASDs as well as their unaffected siblings. The researchers suggest that their method allows the identification of autism risk genes, and demonstrated a high likelihood that mutations in the SCN2A gene impart a risk of developing an ASD.
Autism effects one in every 88 people in the USA, with men being more often affected than women. The causes of autism are thought to be due to a complex interplay of genetic and environmental factors, with many genes involved in determining risk.
This has significant consequences for autism research; Buxbaum believes that “As these genes are further characterized, this will lead to earlier diagnosis and novel drug development. This work is crucial for advancing autism treatment.”
The results from these studies, and others like them, could identify genes involved in the development of ASDs, which would allow advances in both the diagnosis and classification of ASDs and potential therapies for the disorders.
– Written by Alisa Crisp
Sources: Sanders SJ, Murtha MT, Gupta AR et al. De novomutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485(7397), 237–241 (2012); O‘Roak BJ, Vives L, Girirajan S et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485(7397), 246–250 (2012); Neale BM, Kou Y, Liu L et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485(7397), 242–245 (2012).