Novel Polymorphisms in CYP4F2 and VKORC1 May Lead to Adaptations to the Warfarin-Dosage Algorithm
The recent study carried out by researchers from the Lithuanian University of Health Sciences, Lithuania, may enable better targeting of the therapeutic window of warfarin by revealing the influence of polymorphisms in CYP4F2 and VKORC1 on warfarin dosage. The dosage of warfarin, an anticoagulation drug that has been in use for over 50 years, is affected by multiple factors such as patient‘s age, body weight, clinical, environmental and genetic factors. The research group aimed to focus on the genetic factors and, in particular, whether genotypes VKORC1 G3730A and CYP4F2 G1347A may contribute to warfarin dosage due the position of these gene products in the warfarin metabolic pathway.
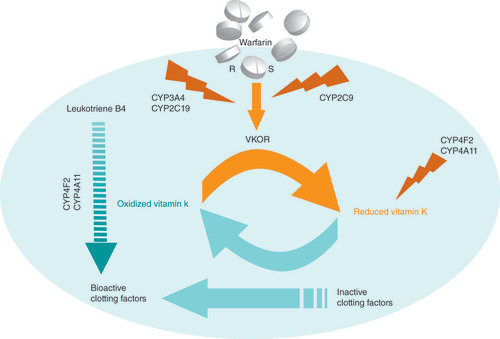
Thus, it stops the production of functional clotting factors. S-warfarin is metabolized by CYP2C9. R-warfarin is metabolized by other highly polymorphic enzymes, such as CYP3A4, CYP2C19 and others. CYP4F2 (also CYP4A11) metabolizes the leukotriene B4 and vitamin K. Leukotriene B4 is a potent inducer of leukocytes and can stimulate the process of inflammation.
R: R-warfarin; S: S-warfarin.
Figure courtesy of Dr Vacis Tatarunas.
The researchers genotyped a total of 307 patients, who had all undergone heart valve surgery. Out of this group, 189 had been receiving warfarin for over 3 months. A multivariate linear regression model showed that during initiation, clinical factors can explain 17% of the warfarin dose variation. Upon addition of CYP2C9 and VKORC1 G1639A genotype to the model the accuracy was approximately doubled, to 32%. The CYP4F2 G1347A genotype was also able to raise the accuracy by a similar amount.
Thus these results show for the first time, that warfarin dosage during induction depends on CYP4F2 (rs2108622) genotype, but it has no effect during stable dosage. In regards to the clinical implications of this work Dr Tatarunas, one of the main authors, informed us that patients with the CYP4F2 A/A genotype “should be anticoagulated with a higher dosage of warfarin during induction after surgery”. By contrast, the effect of VKORC1 (rs7294) genotype remained significant in patients on stabilized dosage.‘
Dr Tatarunas highlighted the significance of the CYP4F2 A1347A genotype, due to its involvement and interaction with many inflammatory mediators and quinine compounds. He outlined that ‘it metabolizes and degrades the leukotriene B4, as well as controlling the metabolism of vitamin K. CYP4F2 also participates in synthesis of 20-hydroxyeicosatetraenoic acid (20-HETE), which has antiplatelet and vasoconstrictor effects.‘ Thus, these results might also have implications in other clinical areas involving inflammation.
Warfarin use comes with a risk, owing to its narrow therapeutic index, therefore improving the warfarin-dosage algorithm is a large focus in this field of research. The results of this study will contribute to the ongoing research striving to adapt the warfarin-dosage algorithm.
Source: Tatarunas V, Lesauskaite V, Benetis R et al. The effect of CYP2C9, VKORC1 and CYP4F2 polymorphism and of clinical factors on warfarin dosage during initiation and long-term treatment after heart valve surgery. J. Thromb. Thrombolysis doi:10.1007/s11239-013-0940-x (2013) (Epub ahead of print).
Genotypes to Phenotypes: Polymorphisms Underlying Clozapine-Induced Obsessive–Compulsive Symptoms in Schizophrenia Patients Uncovered
A research team led by Chen Zhang (Shanghai Jiao Tong University School of Medicine, China) has recently revealed associations between polymorphisms in genes SLC1A1, GRIN2B and GRIK2 and clozapine induced obsessive–compulsive (OC) symptoms in patients with schizophrenia.
The induction of OC symptoms in some schizophrenia patients, upon use of atypical antipsychotics (AAPs), especially clozapine, has been a troublesome phenomenon in psychiatry, with information pertaining to the underlying genetic influences remaining largely unknown.
A logical starting point to investigate these genetic influences was provided by evidence from clinical trials showing that the glutamatergic neurotransmitter within cortical–striatal–thalamocortical circuitry is disrupted in patients with OC disorder (OCD). This led Dr Zhang‘s research team to consider the hypothesis that the mechanism underlying clozapine-induced OC symptoms might share the etiological pathway with OCD.
With this hypothesis in mind, Dr Zhang selected the ‘three most promising candidate OCD-associated genes in glutamatergic transmission‘; SLC1A1, GRIN2B and GRIK2, which previous studies have shown to be associated significantly with OCD. The severity of OC symptoms in 250 clinically stable schizophrenia patients receiving clozapine treatment, were evaluated using the Yale-Brown Obsessive Compulsive Scale (Y-BOCS). Patients were separated into two groups, an OC group and a non-OC group, based on their Y-BOCS score. The three OCD susceptibility polymorphisms, SLC1A1 (rs2228622), GRIN2B (rs890) and GRIK2 (rs1556995) were genotyped and were subsequently tested for an association with clozapine-induced OC symptoms.
The results detected an interaction between SLC1A1 rs2228622A and GRIN2B rs890T alleles, which was associated with OC symptoms. The interaction between these two alleles was found in the significant two-locus gene–gene interaction model and was further confirmed by the significant Y-BOCS score (F6, 137 = 7.650, p < 0.001).
These results strongly suggest that SLC1A1, GRIN2B, and interactions between the two may significantly enhance the risk of OC symptoms. These findings therefore provide preliminary evidence that genes in glutamatergic transmission are associated with clozapine-induced OC symptoms. The results also provide us with a foundation from which the elucidation of the biological and biochemical pathways underpinning clozapine-induced OC symptoms can proceed. Moreover, Dr Zhang emphasizes that clozapine is an ‘excellent tool drug‘ and therefore the implications of this study do not just relate to clozapine use but branch out to cover other AAPs as well. Clozapine, being the first AAP to be developed has been more extensively characterized than the others, which have been created in its shadow and are therefore largely based on clozapine‘s mechanism of action. It is therefore not beyond the bounds of reason to assume that the genotypes identified in this study, may also be associated with adverse events seen with the use of other AAPs. We therefore eagerly await further studies using other AAPs focused on these genotypes
Further replication of these results is required in order to build up and strengthen the links between genotype and phenotype. Once this stage has been reached, then a patient‘s specific pattern of polymorphisms may be used as a predictor for clozapine-induced OC symptoms. Indeed, there may be a foreseeable time in the future where the decisions of psychiatrists, when it comes to prescribing appropriate antipsychotics for schizophrenic patients, are guided by genotyping information. Chen Zhang hopes that this preliminary evidence may be a stepping stone towards the day when drugs are ‘tailor-made for individuals and adapted to each persons own genetic make up.‘
Source: Cai J, Zhang W, Zhang C et al. Influence of polymorphisms in genes SLC1A1, GRIN2B, and GRIK2 on clozapine-induced obsessive-compulsive symptoms. Psychopharmacology (Berl.) doi:10.1007/s00213-013-3137-2 (2013) (Epub ahead of print).
Disease-Free Survival in Acute Myeloid Leukemia Patients Affected by Drug Pathway Polymorphisms
An association between germline mutations in drug pathway genes and disease-free survival (DFS) in adults with acute myeloid leukemia (AML) has been revealed by researchers from the University of California, CA, USA. The impact of tumor somatic mutations on DFS are well known and have been previously reported, however, Dr Andreadis, the main author of the study informs us that this was the first study ‘to systematically examine this (germline mutations and DFS) association in the setting of AML treated with high dosage chemotherapy, which is one of the established standards of care.‘
The DFS for AML patients treated with chemotherapy-based autologous stem cell transplantation varies widely between different patient groups, ranging from 10% to over 70%. This study aimed to explain some of this variation by investigating whether any associations could be found between DFS and polymorphisms in the pharmacokinetic and pharmacodynamic pathway genes of cytarabine, etoposide and busulfan (drugs used for the treatment of patients with first or second remission with a two-step approach to autologous stem cell transplantation).
A total of 154 genetically European patients were included in the study. The researchers genotyped 1659 variants in 42 genes and used the Cox-proportional hazards model to determine any association with DFS. The analysis uncovered a SNP in the intronic region of ABCC3 (rs4148405), which was associated with a significantly shorter DFS in the primary cohort. A SNP in the GSTM1–GSTM5 locus, rs3754446, was also found to be significantly associated with a shorter DFS in all patients.
In the future, the interactions between the SNPs of these drug pathway genes is something that must be investigated. Meanwhile, however, the GSTM1–GSTM5 SNP that was revealed by this study deserves particular attention as it is also associated with busulfan levels in patients. Busulfan is widely regarded as the most active agent in the stem cell transplantation process. Therefore, within this context of known polymorphisms, this result implies that changing the drug dosing and targeting the AUC could have the potential to affect the outcome of AML.
Source: Yee SW, Mefford JA, Andreadis C et al. Impact of polymorphisms in drug pathway genes on disease-free survival in adults with acute myeloid leukemia. Clin. Lung Cancer doi:10.1038/jhg.2013.38 (2013) (Epub ahead of print).
Potential Influence of UGT1A8 Genotype on Osteoporosis and Breast Cancer Treatment Outcome
Raloxifene, an oral selective estrogen receptor modulator, is used extensively in osteoporosis and breast cancer prevention treatment in postmenopausal women. A recent study carried out by researchers from the Penn State Hershey Cancer Institute, PA, USA, has revealed a potential role of the UGT1A8 genotype in raloxifene metabolism, helping to characterize the metabolic process as a whole, something that is crucial for the progression of these research areas.
Raloxifene, typically undergoes glucoronidation to form two products; raloxifene-6-glucuronide and raloxifene-4´-glucuronide As the potency of a drug depends largely on its metabolism, the research team decided to elucidate whether the glucoronidation process may be influenced by functional polymorphism in UGTs.
The researchers assessed raloxifene metabolism using homogenates from HEK293 UGT-overexpressing cell lines. They found that the extrahepatic UGT1A8 enzyme is one of the main enzymes involved in the glucoronidation process. Cell lines overexpressing UGT1A8 variants showed decreased raloxifene glucoronidation activities in vitro. Indeed the formation of raloxifene glucuronides showed significant correlation with UGT1A8*2 variants in human jejunum homogenates. Plasma raloxifene-6- and 4-glucuronide, and total raloxifene glucuronides were increased in raloxifene-treated subjects who were predicted slow metabolizers (UGT1A8*1/*3) versus intermediate metabolizers (UGT1A8*1/*1 or UGT1A8 1/*2) versus fast metabolizers (UGT1A8*2/*2).
These results are indicative of the UGT1A8 genotype impacting raloxifene metabolism. This genotype could therefore potentially affect the overall response to the drug and consequently its influence could extend to treatment outcome as well, making this a worthy genotype for further investigation.
Source: Sun D, Jones NR, Lazarus P. Characterization of raloxifene glucuronidation. Potential role of UGT1A8 genotype on raloxifene metabolism in vivo. Cancer Prev. Res. (Phila.) doi:10.1158/1940-6207.CAPR-12-0448 (2013) (Epub ahead of print).
– All stories written by Katie Lockwood