Abstract
Aim: Our work aimed to designate the optimal DNA source for pharmacogenetic assays, such as the screening for HLA-B*57:01 allele. Materials & methods: A saliva and four buccal swab samples were taken from 104 patients. All the samples were stored at different time and temperature conditions and then genotyped for the HLA-B*57:01 allele by SSP-PCR and classical/capillary electrophoresis. Results: The genotyping analysis reported different performance rates depending on the storage conditions of the samples. Given our results, the buccal swab demonstrated to be more resistant and stable in time with respect to the saliva. Conclusion: Our investigation designates the buccal swab as the optimal DNA source for pharmacogenetic assays in terms of resistance, low infectivity, low-invasiveness and easy sampling, and safe transport in centralized medical centers providing specialized pharmacogenetic tests.
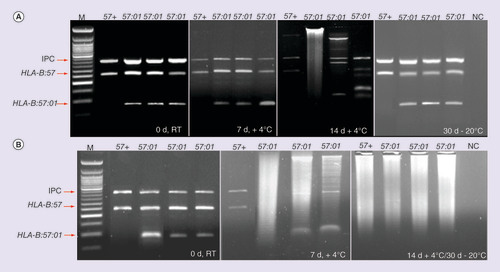
(A) The reported samples refer to the saliva, tested at 0 days (d) at RT, 7d at +4°C, 14d at +4°C and 30d at -20°C. (B) is related to the swab samples, which have been tested at the same time and temperature conditions.
IPC: Internal positive control; M: 50 bp ladder; NC: Negative control.
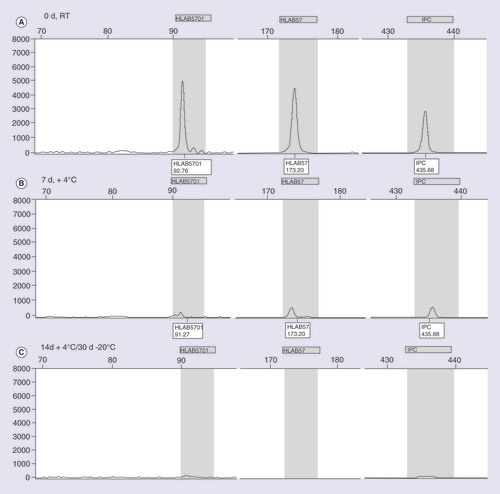
The reported samples refer to the saliva, tested at (A) 0 days (d) at room temperature, (B) 7d at +4°C, and (C) 14d at +4°C and 30d at -20°C.
IPC: Internal positive control.
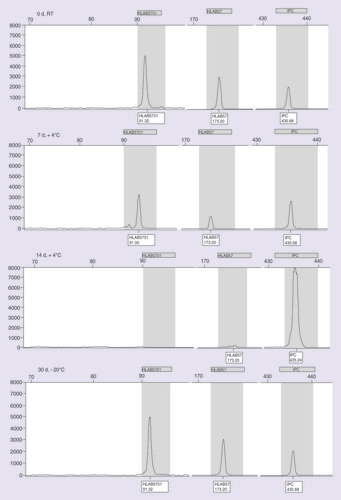
The reported samples refer to the swabs samples, tested at 0 days (d) at RT, 7d at +4°C, 14d at +4°C and 30d at −20°C.
IPC: Internal positive control; RT: Room temperature.
Background
The novel proactive healthcare model is mainly oriented towards the demystification of the disease and the maintenance of the patient welfare, in order to introduce an innovative medicine approach: the 4P Medicine [Citation1]. This healthcare model consists of four specific ‘ingredients’: Prediction, Prevention, Personalization and Participation. The Predictive component concerns the exploitation of the genomic information with the aim of evaluating the individual susceptibility to complex diseases or to the drug response degree. The wide knowledge of the human genome can be further employed for the Prevention of disease conditions and the Personalization of the treatment. The predictive, preventive and personalized medicine are expected to involve the full participation of the patient, who represents the real hinge of the proactive medicine [Citation1,Citation2]. The 4P approach is essentially based on the employment of the human genome variability to improve the individual response to the pharmaceutical treatments and avoid drug adverse reactions. To this purpose, the pharmacogenomics discipline has been developed to promote a safer, more efficient and personalized therapy (i.e., ‘the right drug to the right patient’) [Citation3]. The importance of pharmacogenomics in the clinical practice is provided by the identification of specific genetic polymorphisms (SNPs and copy number variation, [CNV]) that are able to affect the effectiveness and toxicity of the drugs [Citation4]. An excellent example of the pharmacogenetic research application in the clinical practice is the genetic screening for HLA-B*57:01allele in HIV infected patients. The HLA-B*57:01 has been significantly associated with a higher risk of developing hypersensitivity reaction (HSR) after abacavir administration (ABC, one of the most efficient antiretroviral drugs) [Citation5–8]. Economic evaluations of the HLA-B*57:01 screening in the treatment of HIV patients proved to be cost–effective, resulting in a large reduction of the costs required for HSR treatment [Citation9–11]. On the basis of this result, the US FDA and the EMA made the HLA-B*57:01 screening obligatory prior ABC treatment (from the FDA alert released on 24 July 2008: ‘Screening for the HLA-B*5701 allele is recommended for all patients prior to starting abacavir therapy’).
The demand for pharmacogenetic tests providing fast, high-quality, reliable and repeatable data encouraged the centralization of laboratories offering pharmacogenetic analysis. The usefulness of centralized diagnostic services lay on the higher-quality support concerning the analysis and results interpretation, which can be supplied only by specialized centers [Citation12].
The centralization of diagnostic and medical centers providing pharmacogenetic tests has given special emphasis to the infectivity and safe transport of ‘infectious substances.’ The WHO defines these substances as: ‘substances which are known or are reasonably expected to contain pathogens (bacteria, viruses, rickettsiae, parasites, fungi) and other agents such as prions, which can cause disease in humans or animals’ [Citation13]. Over the potential risk of infection, the safe transport of any biological sample should always ensure the preservation of its integrity until the real analysis. In particular packaging, identification and refrigeration should be evaluated before the shipment of the infectious substances [Citation13].
Among the several infectious samples, particular attention has been paid in case of HBV, HCV and HIV presence. To date, the blood is the biological source commonly utilized for molecular analysis. However, in spite of the advantages in terms of quantity and easiness of sampling, the blood is the most infective biological fluid, concerning the risk of transmission of these viruses [Citation14].
This article will focus on the evaluation of less infective HIV samples for diagnostic and pharmacogenetic testing, such as the buccal swab and the saliva. Given the very low infectivity, these biological samples can be safely transported in specialized medical centers, making possible the centralization of specific molecular diagnostic services [Citation12].
One of the most successful centralized analysis is the pharmacogenetic test for HLA-B*57:01 typing.
The possibility of utilizing less infective biological samples have particular importance in the context of the centralization of the diagnostic service, the safe shipment and handling of the tested materials [Citation12,Citation15]. To date different genotyping methods exist for HLA-B*57:01 detection, each one with its own positive and negative aspects. Unfortunately most of them need blood as starting biological sample, except for the SSP-PCR and CE (single specific primer-polymerase chain reaction and capillary electrophoresis) and the Direct-PCR approaches, for which saliva and buccal swab can be employed [Citation15].
The final goal of our work is the evaluation of the integrity and stability of saliva and buccal swab in terms of time and storage, in order to be applied as primary DNA sources for HLA-B*57:01 genotyping.
Materials & methods
The study enrolled 104 HIV-positive patients coming from different infectious disease Italian centers (University of Rome ‘Tor Vergata,’ ‘Tor Vergata’ University Hospital, University of Rome ‘Sapienza,’ ‘Umberto I’ University Hospital, ‘San Giovanni Addolorata’ Hospital, ‘Santo Spirito’ Civil Hospital). Written informed consent for genetic analysis and research analysis was obtained from all the subjects. The study was approved by the institutional ethics committees of all the participating hospitals. Four buccal swabs and a saliva sample were collected in parallel from all 104 cases, thus obtaining a total of 520 samples: 104 of them were saliva, the remaining 416 were buccal swabs (four buccal swabs each patient × 104). Patients were instructed not to eat in the 2 h before the sample collection. Saliva was collected by the patient spitting into the collection tube and subsequently it was split into four single tubes (400 µl). The buccal swabs were obtained by rubbing the cheek for 30 s and placing it in a sterile tube. Four storage conditions were established and tested, in order to determine the sample stability in terms of storage time and temperature. The samples were stored at three different temperatures: Room Temperature (RT), +4°C and -20°C. The saliva single tubes and the buccal swabs were processed and genotyped in four different times: time 0 at RT; time 7 and 14 days at +4°C; time 30 days at -20°C.
DNA was extracted from saliva and buccal swabs with the DNA IQ™ System-Kit (Promega, WI, USA) according to the manufacturer’s instructions. DNA IQ™ System is a manual method for DNA extraction, which is based on the utilization of magnetic beads to isolate the DNA from the biological sample and special buffers for the DNA extraction, purification and elution.
The Nanodrop 1000 spectrophotometer (NanoDrop Technologies, DE, USA) was used to evaluate the concentration and the purity (Optical Density, OD) of the extracted DNA, assessing the standard UV absorbance at 260 nm, the ratios of absorbance at 260 nm/280 nm (A260/280) and at 260 nm/230 nm (A260/230). The acceptable ranges for OD values of Saliva and buccal swab samples were fixed to 1.6–2.0 for A260/280 and 1.5–2.0 for A260/230.
The HLA-B*57:01 pharmacogenetic test was utilized to test the DNA sources (saliva and buccal swabs) in terms of quality and resistance. In order to determine which biological sample allows higher quality DNA for genotyping analysis, two molecular assays for HLA-B*57:01 detection were performed: SSP-PCR & Classical Electrophoresis and SSP-PCR & Capillary Electrophoresis. The quality and the stability of the DNA was evaluated by separation of the amplicons with classical electrophoresis on 2,5% agarose gel and Capillary electrophoresis on ABI3130 XL automated sequencer (Applied Biosystems).
SSP-PCR & Classical Electrophoresis consists of a multiplex-PCR for the amplification of HLA-B*57 (~260 bp), HLA-B*57:01 (94 bp) and a control region (~400 bp, internal control). The PCR was performed according to the protocol developed by Mallal et al. [Citation16]. The presence of the three amplified regions was proved by observation of the intensity and the size of the bands on the gel after electrophoretic separation (120 V for 40 min). In particular, the correct size of the amplicons was determined by comparison with a 50 bp Ladder (BioLabs).
SSP-PCR & Capillary Electrophoresis is characterized by the amplification of HLA-B*57 (~175 bp), HLA-B*57:01 (92 bp) and a control region (~400 bp, internal control). The PCR was performed according to the protocol developed by Giardina et al. [Citation17]. The presence of HLA-B*57:01 allele was evaluated on the basis of the fluorescence and the peak amplitude of the PCR products separated by capillary electrophoresis. The collection and the analysis of the data obtained by capillary electrophoresis were assessed by GeneMapper® software (Applied Biosystems).
Results
All the 520 samples have been genotyped by SSP-PCR and classical/capillary electrophoresis, reporting different performances in terms of storage temperature and time conditions. The intensity of the bands on the agarose gel and the fluorescence unit on the electropherograms were descriptive of the quality and integrity of the extracted DNA. As expected, the degradation of the extracted DNA was recognizable by the smears on the agarose gel and the absence of fluorescence peaks on the electropherograms.
All the samples were processed and genotyped after conservation at different times and temperature conditions: time 0 at RT; time 7 and 14 days at +4°C; time 30 days at -20°C.
Samples processing consisted of the DNA extraction, followed by the measurement of OD values. Concerning the saliva, we observed good OD values for fresh samples (time 0, A260/280 ∼1.8-A260/230 ∼1.7), in contrast to the progressively lower values recorded with the stored samples (time 7, A260/280 ∼1.6 - A260/230 ∼1.5; time 14, A260/280 ∼1.2 - A260/230 ∼1.0; time 30, A260/280 ∼1 - A260/230 ∼0.5). On the other hand, buccal swabs showed good OD values for fresh samples (time 0, A260/280 ∼1.9 - A260/230 ∼1.9) and processed after 30 days at -20°C (time 0, A260/280 ∼1.9 - A260/230 ∼1.9). For the buccal swabs stored at time 7 and 14 at +4°C, decreasing OD values were recorded (time 7, A260/280 ∼1.7 - A260/230 ∼1.6; time 14, A260/280 ∼1.0 - A260/230 ∼0.8).
According to the workflow previously defined, a saliva sample and a buccal swab per patient were firstly typed at time 0. The extracted DNA evaluated by SSP-PCR and classical/capillary electrophoresis showed high quality in terms of concordance of the resulting genotype and reproducibility of the molecular assay. In fact, 100% of the analyzed saliva and swab samples gave reliable and accurate results (positivity/negativity for HLA-B*57:01).
The second phase of the study consisted of the processing of the saliva and the buccal swab after storage for 7 days at +4°C. Concerning the saliva samples, the extracted DNA demonstrated low quality, degradation and nonreproducibility. The genotyping results visualized on the agarose gel were not reliable due to the presence of smears and weak amplicons bands. The electropherograms referred to the same samples showed a reduction in terms of fluorescence intensity and resolution of the peaks. The 12% of the saliva samples were correctly genotyped (positivity/negativity for HLA-B*57:01). On the other hand, the analysis of the buccal swabs reported higher quality and reproducibility rates, nearly matching with those obtained at time 0. In this case, the genotype of the 80% of the patients was accurately determined (positivity/negativity for HLA-B*57:01).
The +4°C temperature condition has been further evaluated testing the stability of the samples after the storage for 14 days. As expected, the DNA from saliva samples was entirely disrupted, resulting in a complete absence of detectable amplification by classical and capillary electrophoresis. None of the 104 saliva samples gave reliable results. Concerning the buccal swabs, a partial degradation and misinterpretation of the results have been observed in both genotyping methods. In this case only the 15% of swab samples were properly genotyped (positivity/negativity for HLA-B*57:01).
The last tested storage and temperature conditions are related to the samples genotyping after 30 days at -20°C. While the saliva showed the same disruption signs of the previous storage condition, the buccal swabs proved to be resistant and undamaged. The extracted DNA allows the accurate and reliable analysis and interpretation by SSP-PCR and classical/capillary electrophoresis, as shown by the successful genotyping of the 100% of the amplified swab samples.
– depict the results (SSP-PCR and classical/capillary electrophoresis, respectively) exemplifying the different storage and temperature conditions related to the saliva samples. In A it is possible to notice the progressive degradation of the saliva samples on agarose gel from time 0 until 30 days. The reduction of peak resolution and fluorescence intensity on electropherograms, instead, are shown in . The genotyping of the swab samples showed to be successful at time 0 at RT; 7 days at +4°C and 30 days at -20°C condition, as illustrated in B–.
Discussion
The Pharmacogenomic tests represent one of the latest fields of application of the genetic polymorphisms in the clinical practice. The constant progress of the pharmacogenomics paves the path for the implementation of the personalized medicine approach according to the novel proactive healthcare model [Citation3].
Our work concerned the HLA-B*57:01 allele as it represents the best qualified pharmacogenetic biomarker to be utilized in the clinical practice. It is well-known that HLA-B*57:01-positive subjects cannot be treated with abacavir in case of HIV infection, because of the higher risk of HSR development. Abacavir is antiretroviral drug, utilized as a second-line treatment of HIV infection, especially in pediatric patients [Citation18]. It is characterized by a good oral bioavailability and low toxicity profile, although it is responsible of HSR in the 5% of the treated patients. The adverse reaction has been strongly associated with the presence of HLA-B*57:01allele, thus restricting its prescription. The clinical advantages of HLA-B*57:01 pharmacogenetic screening before ABC administration are the possibility to prevent adverse reaction and ensure a more personalized, effective and safer treatment [Citation4]. Given these data, the development of accurate molecular assays for HLA-B*57:01 detection are of crucial importance to ensure the necessary expertise and promote the implementation of the pharmacogenetic test. In this context, the selection of the optimal biological sample utilized as source of DNA has a strong relevance, in terms of potential infectivity, preservation of the sample integrity and safe transport. Concerning the potential infectivity of the DNA sources, the employment of biological materials alternative to the blood for HLA-B*57:01 screening, may be of help in avoiding the risks of HIV exposure and infection. On this subject, our experience in the field of pharmacogenetics demonstrated that saliva and buccal swab are two of the best qualified DNA sources in terms of low infectivity, manageability and safe transport (in view of the potential development of a centralized analysis in specialized laboratories) [Citation19,Citation20]. In this context, the aim of our work concerned the ‘nomination’ of the more stable and reliable DNA source between the saliva and the buccal swab. We observed that at time 0, both the biological samples provide similar high quality results, while their performance rates change at longer storage conditions. In particular, the saliva stored at +4°C for 7 days (or more) and at -20°C for 30 days shows loss of integrity, compromising the genotyping analysis. In contrast, the buccal swab can be stored for up to 7 days at +4°C or 30 days (and more) at -20°C, preserving its integrity and providing accurate genotyping. Unfortunately, the swab sample stored for a time longer than 7 days at +4°C (14 days or longer) demonstrated to be disruptive and inadequate for the analysis.
We suppose that the higher vulnerability of saliva may depend on the presence of some lytic enzymes and general PCR inhibitors that compromise the accessibility to DNA and the overall amplification yield. Moreover, the freezing of saliva may be responsible of structural changes which result to be disruptive for the maintenance of the sample integrity. Therefore, the wrong conservation of the saliva samples induces the rapid degradation of DNA, undermining the final outcome of the pharmacogenetic test and forcing the operator to contact again the patient and repeat the sampling procedure.
Given our results, the buccal swab showed higher resistance and stability for a longer time with respect to the saliva. In fact, the employment of buccal swab as DNA source allows to bypass all the ‘limits’ due to the composition of saliva, especially the greater amount of lytic enzymes and PCR inhibitors. In contrast, the sampling procedure and the buccal swab in itself facilitate the collection of cleaner samples, containing a larger amount of epithelial cells suitable for the DNA extraction. Moreover, it is important to consider the attitude of the patient in front of the sampling of the buccal swab or the saliva, preferring the swab sample to the saliva sampling procedure.
Conclusion
The introduction of pharmacogenetic test in the clinical practice and the need for DNA sources which are alternative to the blood, may be advantageous not only for the patient but even for the National Health Service. In this context the buccal swab represents an excellent solution concerning the costs required for sampling materials (needle, blood collection tube, alcohol, tourniquet), sample transport, safety for the operators and invasiveness of the sampling procedure.
Our investigation designates the buccal swab as the optimal DNA source for pharmacogenetic and molecular assays, such as the screening for HLA-B*57:01 genotyping. In fact the buccal swab showed to be more stable, resistant and less subject to PCR inhibitors. In addition, the low infectivity, the comfort and the easiness of sampling and transport, make the buccal swab the suitable candidate to promote the centralization of molecular diagnostic services and the development of new pharmacogenetic assays. Moreover the low-invasiveness and easiness of the sampling procedure support the participation and the involvement of the patient, who is the active player along his own diagnostic-therapeutic pathway. Therefore, the development of centralized pharmacogenetic analysis utilizing the buccal swab as DNA source fall within the main principles characterizing the proactive healthcare model. In fact, it is finally focused on the improvement of the patient care, by offering safe, effective and personalized medical services. In conclusion, the new medicine concept (4P) should rely on the active contribution of patients, operators and general consumers in order to promote the maintenance of human welfare and provide noninvasive, comfortable pharmacogenetic tools for a personalized treatment and diagnosis course.
The selection of the primary source of DNA is of crucial importance to ensure high-quality and reliable results of the pharmacogenetic tests.
The final goal of our work was the evaluation of the integrity and stability of saliva and buccal swab in terms of time and storage, in order to be applied as primary DNA sources for pharmacogenetic testing.
Saliva demonstrated to be more subject to PCR inhibitors, time and temperature storage conditions (<7 days at +4°C or -20°C), which undermine the sample integrity and compromise the final outcome of the pharmacogenetic test.
The buccal swab showed to be more stable, resistant and less subject to PCR inhibitors, time and temperature storage conditions (till 7 days at +4°C or for long time at -20°C).
The buccal swab is one the best qualified DNA source, in terms of low-infectivity, manageability, safe transport and costs required for the sampling procedure.
The employment of DNA sources which are alternative to the blood, promotes the centralization of laboratories in order to provide higher quality and specialized support concerning the analysis and the interpretation of the pharmacogenetic data.
The availability of resistant, manageable and inexpensive DNA sources encourage the development of new pharmacogenetic assays, which will be able to provide more and more effective, safe and personalized treatment.
Acknowledgements
The authors thank ViiV Healthcare s.r.l., Verona, Italy, for the support provided in the identification of clinical centers willing to collaborate to the present work and in monitoring progression of activity in those centers. No economic support was provided by ViiV Healthcare.
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
- Hood L , FloresM . A personal view on systems medicine and the emergence of proactive P4 medicine: predictive, preventive, personalized and participatory . N. Biotechnol.29 ( 6 ), 613 – 624 ( 2012 ).
- Hood L . Systems biology and p4 medicine: past, present, and future . Rambam Maimonides Med.4 ( 2 ), e0012 ( 2013 ).
- Cohen JP . Overcoming regulatory and economic challenges facing pharmacogenomics . N. Biotechnol.29 ( 6 ), 751 – 756 ( 2012 ).
- Ma Q , LuAY . Pharmacogenetics, pharmacogenomics, and individualized medicine . Pharmacol. Rev.63 ( 2 ), 437 – 459 ( 2011 ).
- Phillips E , MallalS . Successful translation of pharmacogenetics into the clinic: the abacavir example . Mol. Diagn. Ther.13 ( 1 ), 1 – 9 ( 2009 ).
- Hughes S , HughesA , BrothersC , SpreenW , ThorbornD . CNA106030 Study Team . PREDICT-1(CNA106030): the first powered, prospective trial of pharmacogenetic screening to reduce drug adverse events . Pharm. Stat.7 ( 2 ), 121 – 129 ( 2008 ).
- Mallal S , PhillipsE , CarosiGet al. PREDICT-1 Study Team . HLA-B*5701 screening for hypersensitivity to abacavir . N. Engl. J. Med.358 ( 6 ), 568 – 579 ( 2008 ).
- Saag M , BaluR , PhillipsEet al. Study of Hypersensitivity to Abacavir and Pharmacogenetic Evaluation Study Team . High sensitivity of human leukocyte antigen-b*5701 as a marker for immunologically confirmed abacavir hypersensitivity in white and black patients . Clin. Infect. Dis.46 ( 7 ), 1111 – 1118 ( 2008 ).
- Schackman BR , ScottCA , WalenskyRP , LosinaE , FreedbergKA , SaxPE . The cost–effectiveness of HLA-B*5701 genetic screening to guide initial antiretroviral therapy for HIV . AIDS22 , 2025 – 2033 ( 2008 ).
- Nieves Calatrava D , Calle-Martín OdeL , Iribarren-LoyarteJAet al. Cost–effectiveness analysis of HLA-B*5701 typing in the prevention of hypersensitivity to abacavir in HIV+ patients in Spain . Enferm. Infec. Microbiol. Clin.28 ( 9 ), 590 – 595 ( 2010 ).
- Kauf TL , FarkouhRA , EarnshawSR , WatsonME , MaroudasP , ChambersMG . Economic efficiency of genetic screening to inform the use of abacavir sulfate in the treatment of HIV . Pharmacoeconomics28 ( 11 ), 1025 – 1039 ( 2010 ).
- Stocchi L , CascellaR , ZampattiS , PirazzoliA , NovelliG , GiardinaE . The pharmacogenomic HLA biomarker associated to adverse abacavir reactions: comparative analysis of different genotyping methods . Curr. Genomics13 ( 4 ), 314 – 320 ( 2012 ).
- WHO Guidance on Regulations for the Transport of Infectious Substances 2013–2014, WHO/HSE/GCR/12 (2012) .
- Dianzani F , AntonelliG , CapobianchiMR , DoleiA . Prophylaxis and therapy of viral infections/human retrovirus . In : Manuale di Virologia Medica (Volume 10 and 18).McGraw-Hill , Italy . 117 – 119 / 233 – 253 ( 2008 ).
- Cascella R , StrafellaC , RagazzoMet al. Direct PCR: a new pharmacogenetic approach for the inexpensive testing of HLA-B*57:01 . Pharmacogenomics J.1 – 5 ( 2014 ).
- Martin AM , NolanD , MallalS . HLA-B*57.01 typing by sequence-specific amplification: validation and comparison with sequence-based typing . Tissue Antigens65 ( 6 ), 571 – 574 ( 2005 ).
- Giardina E , StocchiL , CuzzolaVet al. A fluorescence-based sequence-specific primer PCR for the screening of HLA-B(*)57:01 . Electrophoresis21 , 3525 – 3530 ( 2010 ).
- WHO . Antiretroviral medicines in low- and middle-income countries: forecasts of global and regional demand for 2012–2015 . AIDS Medicines and diagnostics service; HIV/AIDS Programme ( 2013 ).
- Giardina E , PietrangeliI , MartoneCet al. Whole genome amplification and real-time PCR in forensic casework . BMC Genomics ( 10 ), 159 ( 2009 ).
- Badulli C , SbarsiI , Di GiorgioDet al. A new approach to safely type for HLA the HIV infected people eligible to abacavir therapy: saliva or buccal swab as reliable DNA sources . Clin. Chim. Acta412 , 1995 – 1998 ( 2011 ).
