Abstract
Aim: To explore the association between miR-938rs2505901 T>C polymorphism and Hirschsprung disease (HSCR) risk in Chinese children. Materials & Methods: We conducted a case–control study in a Chinese population with 1381 cases and 1457 controls. The associated correlation strengths were assessed by adjusted odds ratios (AORs) and 95% CIs. Results: The results revealed that the rs2505901 TC and rs2505901 TC/CC genotype were related to an increased HSCR risk compared to the risk contributed by the rs2505901 TT genotype. A stratification analysis showed that the rs2505901 TC/CC genotype promoted the progression of HSCR more significantly in patients with the short-segment HSCR subtype. Conclusion: Our study indicated that miR-938rs2505901 T>C polymorphism is significantly associated with HSCR risk in Chinese children. This result needs to be confirmed with well-designed studies.
Lay abstract
Hirschsprung disease (HSCR) is the most common intestinal disorder in infants. Genetic defects play important roles in the occurrence and development of HSCR. This case–control study was performed by collecting data from 1381 cases and 1457 controls to evaluate the association between the miR-938rs2505901 T>C genetic defect and HSCR risk. The results showed that allele rs2505901 C may be a risk factor. The rs2505901 TC or rs2505901 TC/CC genotype was related to an increased HSCR risk compared to the risk contributed by the rs2505901 TT genotype. An expression quantitative trait locus (eQTL) analysis indicated that allele rs2505901 C is linked with high levels of miR-938. Our study may provide a target and a valuable reference for the clinical treatment, classification and prognosis of HSCR.
Hirschsprung disease (HSCR), also known as aganglionic megacolon, is the most frequent developmental disorder of the enteric nervous system (ENS) in children and it is characterized by an absence of intramural ganglion cells in the myenteric and submucosal plexuses of the intestinal tract, leading to tonic contraction of the influenced segment, bowel obstruction and massive expansion of the proximal gut [Citation1–4]. Based on the length of the aganglionic segment, patients with HSCR can be anatomically divided into three subtypes: short-segment (S-HSCR, ∼80% of cases), in which the affected segment includes the rectum and a short fraction of the colon; long-segment (L-HSCR, ∼15%), which affects a longer range of the colon; and total colonic aganglionosis (TCA, ∼5%) [Citation5,Citation6]. The morbidity of patients with this disease shows significant racial variation among different ethnic groups; for example, the incidence of HSCR among Hispanic, Caucasian–American, African–American and Asian populations was reported to be approximately 10, 15, 21 and 28 per 100,000 live births, respectively, and the incidence rate was reported to be highest in Asia [Citation1,Citation4]. HSCR shows sex-dependent penetrance with a ratio of 4:1 male:female [Citation7] and presents a non-Mendelian malformation with low penetrance [Citation8]. Most HSCR cases are sporadic, and only 5–20% of cases are familial forms. However, the hereditary cause for these diverse phenotypes, such as the length of the aganglionic segment, male preponderance, high recurrence and associated phenotypes among posterity, and so on, remain largely unclear.
HSCR arises when enteric precursor cells (EPCs), which develop from neural crest cells (NCCs), fail to fully colonize the gut. This defective gut colonization is attributed to the abnormities in EPC proliferation, differentiation, survival and migration during ENS development [Citation6]. Therefore, various genes and pathways involved in these cellular events may affect the genesis and progression of HSCR. To date, at least 15 susceptible genes have been found to be associated with the development of HSCR, and they encode proteins that are crucial for the development of enteric ganglia [Citation9]. These genes function through two pathways. The genes RET, GFRα1, GDNF, NTN and PSPN are expressed in the RET pathway; EDNRB, EDN3 and ECE-1 are expressed in the EDNRB pathway and SOX10, ZFXH1B and PHOX2B are expressed in both the RET and/or EDNRB pathways [Citation4,Citation10]. Additionally, molecular anomalies associated with HSCR have been revealed in the last few years, including copy number variation (CNV) [Citation11], polymorphisms in the 3′ untranslated region (UTR) [Citation12], gene–gene interaction [Citation13] and microRNA interactions [Citation14].
MicroRNAs (miRNAs) are small (21–25 nt), noncoding, single-stranded RNA molecules that are crucial for the posttranscriptional regulation of gene expression. These RNA species function as negative regulators by binding to the 3′ UTRs of target mRNAs, depending on the specific sequence and resulting in mRNA transcript degradation and repression of translation [Citation15,Citation16]. One single miRNA can bind to and regulate numerous targets, and more than one-third of human messenger RNAs (mRNAs) are regulated by various miRNAs that are required for normal cellular functions [Citation17,Citation18]. Previous studies have demonstrated that miRNAs play key roles in maintaining energy homeostasis, particularly in the regulation of intestinal homeostasis, the cell cycle, cell proliferation, cell apoptosis, cell migration and neuronal function [Citation19,Citation20]. Dysregulated miRNAs have been shown to be involved in HSCR development, including miR-206 [Citation21], miR-192/215 [Citation22], miR-939 [Citation23], miR-770-5p [Citation24], and miR-214 [Citation25], which suppress cell proliferation and migration. Thus, miRNAs are considered susceptibility genes that contribute to HSCR development, and many have been studied. Among these miRNAs, miR-938 has been studied extensively in cancer. It was reported that miR-938 is expressed at a low level in colon cancer patients, a trend most prominent from the early to the late tumor-lymph node metastatic stages, and this information can be used to distinguish non-invasive healthy individuals from colon cancer patients [Citation26]. However, another study by Li et al. showed that miR-938 was upregulated in colorectal cancer and promoted colon cancer cell proliferation by inhibiting PHLPP2 expression [Citation27]. It is possible that miR-938 exerts diverse functions in the progression of cancer by regulating different downstream targets. Several studies have shown miR-938 polymorphisms associated with complex human diseases in addition to cancer, such as human Type 1 diabetes [Citation28], primary ovarian insufficiency [Citation29], gastric cancer [Citation30,Citation31] and recurrent implantation failure [Citation32]. However, no research has been reported on the associations of miR-938 polymorphisms with HSCR risk; thus, we conducted this case–control study to evaluate whether there is an association in the Chinese population.
Materials & methods
Study population
In total, 1470 HSCR patients were histopathologically diagnosed by histological examination of biopsy specimens that showed a lack of enteric ganglia [Citation33], and data on 1473 geographically and ethnically matched control subjects without an HSCR history or a neurological disorder were collected between 2000 and 2015. All the subjects were recruited from Guangzhou Women and Children’s Medical Center. However, only the 1381 cases and 1457 controls were successfully genotyped. The detailed demographic information of the study participants is summarized in Supplementary Table 1. Written informed consent was signed by the guardians of all participants. The study protocol was coincident with the ethical guidelines of the 1975 Declaration of Helsinki and authorized by the institutional review board of Guangzhou Women and Children’s Medical Center (ethics approval no. 201943800). The authors are accountable for every aspect of the work, assuring that questions about the accuracy or integrity of any portion of the work are appropriately investigated and resolved.
Polymorphism selection & genotyping
In this study, one potentially functional polymorphism in miR-938 (rs2505901 T>C) was eventually chosen, as we describe above [Citation34]. The polymorphism is located in the pri-miR-938 coding region and may affect the maturity and expression of miR-938. In brief, we searched for potentially functional SNP candidates located in the 5′-flanking region, 5′ untranslated region, 3′ untranslated region and exon of miR-938 gene. For genotyping, genomic DNA was extracted from venous blood or paraffin-embedded tissue specimens. Then, genotyping was conducted for selected polymorphisms in purified DNA samples by the classical TaqMan PCR method [Citation35–37]. To ensure the authenticity of the data, 10% of the DNA specimens were selected at random for a second analysis. A 100% concordance rate was obtained in the repeated genotyping.
eQTL analysis
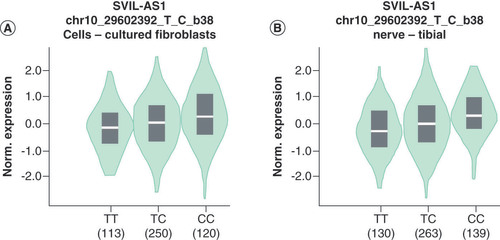
eQTLs are loci or markers in genomes that are associated with gene expression. The genotype-tissue expression (GTEx) project is devoted to exploring the association between genetic variation and gene expression in normal human tissues. In this study, we evaluated the relationship between miR-938rs2505901 T>C variants and miR-938 expression in cell-cultured fibroblasts and nerve-tibial tissues by eQTL analysis using the GTEx portal. The details of the aim, design and data analysis of the study were described in a previous study [Citation38].
Statistical analysis
The goodness-of-fit χ2 test was used to assess whether the selected polymorphism was in HWE among the control subjects. Two-sided χ2 test was applied to calculate the distributions of demographics and allele frequencies among all the cases and controls. The correlation intensity between the selected miR-938 polymorphism and HSCR susceptibility was confirmed by calculating the odds ratios (ORs) and 95% CIs. In addition, we performed a stratification analysis according to the aganglionic segment length in the gut. All statistical analyses were performed in SAS software version 9.4 (SAS Institute, NC, USA). The results were deemed to be statistically significant when the p-values <0.05.
Results
Associations between miR-938 polymorphism and HSCR susceptibility
In general, 1381 cases and 1457 control genotypes were successfully identified in this case–control study. As shown in , the genotype frequency distribution of the selected miR-938rs2505901 T>C polymorphism aligned with Hardy–Weinberg equilibrium (HWE) among the controls (HWE = 0.714). We found that miR-938rs2505901 T>C was related to HSCR risk. Specifically, miR-938rs2505901 TC genotype increased HSCR susceptibility compared to the effect of the rs2505901 TT genotype (AOR = 1.22; 95% CI = 1.04–1.43; p =0.016), and the rs2505901 CC genotype enhanced the HSCR risk more intensely than the rs2505901 TT genotype, but the difference was not statistically significant (AOR = 1.35; 95% CI = 0.97–1.87; p = 0.078). However, in a dominant model, the rs2505901 TC/CC genotype also increased the risk of HSCR more intensely than the rs2505901 TT genotype (AOR = 1.24; 95% CI = 1.06–1.44; p = 0.007).
Table 1. Association between miR-938 rs2505901 T>C polymorphism and Hirschsprung disease susceptibility.
Stratification analysis of miR-938 polymorphism-associated risk according to HSCR subtype
To explore whether the selected miR-938 polymorphism exerts a diverse risk effect on HSCR among different subgroups, a stratification analysis was performed based on the segment length of aganglionic segment. The HSCR subjects were classified into SHSCR, LHSCR and TCA subtypes (). The data revealed that association of miR-938rs2505901 T>C polymorphism with the HSCR susceptibility was limited to the SHSCR subtype, that is, the rs2505901 TC/CC genotype significantly increased SHSCR susceptibility compared with the rs2505901 TT genotype (AOR = 1.31; 95% CI = 1.11–1.55; p = 0.002). However, in the other two HSCR subtypes, LHSCR and TCA, no significant correlation was found.
Table 2. Stratification analysis for the association between miR-938rs2505901 T>C polymorphism and Hirschsprung disease susceptibility (by subtype).
Genotype-based miRNA expression analysis
These results indicated that miR-938rs2505901 T>C polymorphism was related to HSCR risk. However, how do the rs2505901 T>C variants affect HSCR risk? Does the rs2505901 T>C polymorphism affect the expression of miR-938? To assess this hypothesis, we explored the biological effects of three variant genotypes of miR-938 expression by eQTL analysis using the GTEx portal (). The results showed that the rs2505901 TC genotype was associated with an increased expression of miR-938 compared with the rs2505901 TT genotype. The rs2505901 CC genotype upregulated miR-938 expression to a greater extent than the other genotypes. This genotype-based increase in the expression of miR-938 may contribute to HSCR susceptibility.
Discussion
To study the effect of miR-938 polymorphism on HSCR risk, we performed a case–control study with a sample of the Chinese population. We discovered that miR-938rs2505901 T>C polymorphism was significantly related to miR-938 expression, and that this polymorphism was significantly associated with HSCR susceptibility.
Because of their ability to extensively regulate post-transcription gene expression by binding to the 3′ UTR of targeted mRNA, miRNAs are considered as important in disease susceptibility. Aberrant expression of miRNAs has been widely reported in various pathological processes and diseases, including cancers [Citation39] and diabetes [Citation40]. Previous studies have shown that miRNAs are involved in the development of HSCR [Citation14,Citation41]. Diverse mechanisms, such as chromosomal rearrangements, gene amplification or deletion and epigenetic regulation, can result in abnormal miRNA expression [Citation42]. Single-nucleotide polymorphisms represent a common genetic variation that changes the expression and structure of various genes, leading to different levels of disease susceptibility [Citation35]. Polymorphisms in miRNA genes may transform the nature of miRNAs, such as their expression and/or maturation rate [Citation43]. Sequence aberrance near processing sites may have a profound influence on miRNA biogenesis and sequence variation in the mature miRNA, particularly when the anomaly is in the seed sequence, which would change the transcription of the primary miRNA transcript, affecting the interaction between miRNA and mRNA and ultimately causing dysregulated expression of hundreds of target genes [Citation44,Citation45]. Polymorphisms of pre-miRNAs were first reported by Iwai et al. in 2005; this group identified a C-to-A substitution in the mature miR-30c-2 sequence, which may have altered target selection and thus led to profound biological effects [Citation46]. Since this discovery, numerous miRNA polymorphism association studies have been carried out. In 2009, Xu et al. found that miR-196a genetic variant rs11614913 CC can influence mature miR-196a expression and shows decreased binding to the HOXB8 gene, playing a key role in sporadic congenital heart disease susceptibility [Citation47]. More recently, He et al. found two neuroblastoma susceptibility-associated miRNA SNPs: miR-34b/crs4938723 T>C and miR-218rs11134527 A >G, demonstrating that these two polymorphisms exert a protective role against neuroblastoma [Citation48]. A previous study reported that miR-938 was involved in the TGF-β signalling pathway [Citation39]. TGF-β is known as a crucial regulator of cell proliferation and differentiation [Citation49,Citation50]. In 2017, Li et al. showed that miR-938 functions as an oncogene that promotes colon cancer cell proliferation by directly binding to the 3′ UTR of PHLPP2 and suppressing PHLPP2 expression [Citation27]. More recently, Qian et al. proved that miR-938 was upregulated in lung adenocarcinoma tissue and this aberrantly expressed miR-938 promoted lung adenocarcinoma by targeting RBM5 [Citation51]. Furthermore, in 2012, one study conducted by Arisawa et al. suggested that miR-938rs2505901 T>C polymorphism was related to gastric cancer susceptibility [Citation31]. In the present study, we investigated, for the first time, the association between miR-938 polymorphism and HSCR susceptibility. Our results showed that miR-938rs2505901 T>C polymorphism was associated with HSCR risk and that the rs2505901 the TC/CC genotype was associated with increased HSCR susceptibility. Perhaps the rs2505901 C allele functions as a risk allele that contributes to HSCR susceptibility. A genotype-based miRNA expression analysis revealed that the rs2505901 TC/CC genotype was associated with increased expression of miR-938 and that the rs2505901 C allele may contribute to this upregulated expression. Thus, this increased susceptibility may be attributed to the rs2505901 TC/CC genotype-associated abnormal expression of miR-938. Furthermore, we found that the risk effect of the rs2505901 T>C polymorphism on HSCR susceptibility was concentrated in the early stage of HSCR (SHSCR), which indicates that this polymorphism may play a pivotal role in the early progression of HSCR, and that it may be a target for the early prevention and treatment of HSCR. However, the underlying mechanisms should be further investigated. From a clinical perspective, our study may provide a target for the clinical treatment of HSCR. The rs2505901 T>C variant may be a valuable reference for the classification and prognosis of HSCR.
In the present study, several limitations are notable. First, only one polymorphism in miR-938 was investigated, and sometimes the effect of this polymorphism was too weak to determine its function. Second, the subjects involved in this study were all from the same area of China; therefore, the conclusions drawn from this current study may not be generalizable to people living in other regions and to those with other ethnicities. Finally, to explore the potential mechanisms of the risk effect, other molecular, biological and functional experiments should be performed.
Conclusion
In conclusion, our present results suggest that the genetic variant of miR-938rs2505901 was associated with miR-938 expression and affected HCSR susceptibility. Our findings should be further confirmed by choreographed multi-center studies with a larger sample size. In addition, mechanism research should be covered to illuminate the underlying mechanisms by which miR-938 polymorphisms influence HCSR susceptibility.
In this study, we aimed to explore the association between miR-938rs2505901 T>C polymorphism and Hirschsprung disease (HSCR) risk.
Our study results show that the rs2505901 TC and rs2505901 TC/CC genotypes are related to increased HSCR risk and also associated with upregulated miR-938 expression.
In conclusion, our results suggest that miR-938rs2505901 T>C polymorphism is associated with miR-938 expression and affects HSCR susceptibility.
Our findings should be confirmed with coordinated multicentre studies and larger sample sizes.
In addition, mechanistic research should be performed to illuminate the underlying mechanisms by which miR-938 polymorphisms influence HSCR susceptibility.
Author contributions
Conception and design: Wei Zhong and Qiang Wu. Administrative support: Wei Zhong. Provision of study materials or patients: Jun Zhong, Yi Zheng, Xiaoli Xie, Qiuming He and Wei Zhong. Collection and assembly of data: Jun Zhong, Jiabin Liu and Qiang Wu. Data analysis and interpretation: Jun Zhong. Manuscript writing: All authors.
Ethical conduct of research
The study protocol was coincident with the ethical guidelines of the 1975 Declaration of Helsinki and authorized by the institutional review board of Guangzhou Women and Children’s Medical Center (ethics approval no. 201943800). Written informed consent was signed by the guardians of all participants.
Supplemental Table 1
Download MS Word (84.5 KB)Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.tandfonline.com/doi/suppl/10.2217/pme-2021-0001
Financial & competing interests disclosure
This study was funded by grants from the Natural Science Foundation of Guangdong Province, China (no. 2019A1515010971), and Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease (no. 2019B030301004). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or any financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
English language editing was provided by American Journal Experts (AJE).
Data availability
The data used to support the findings of this study are available upon request.
Additional information
Funding
References
- Amiel J , Sproat-EmisonE , Garcia-BarceloMet al. Hirschsprung disease, associated syndromes and genetics: a review. J. Med. Genet.45(1), 1–14 (2008).
- Burzynski G , ShepherdIT , EnomotoH. Genetic model system studies of the development of the enteric nervous system, gut motility and Hirschsprung’s disease. Neurogastroenterol. Motil.21(2), 113–127 (2009).
- Barlow AJ , DixonJ , DixonMJ , TrainorPA. Balancing neural crest cell intrinsic processes with those of the microenvironment in Tcof1 haploinsufficient mice enables complete enteric nervous system formation. Hum. Mol. Genet.21(8), 1782–1793 (2012).
- Tam PK , Garcia-BarceloM. Genetic basis of Hirschsprung’s disease. Pediatr. Surg. Int.25(7), 543–558 (2009).
- Tang CS , ChengG , SoMTet al. Genome-wide copy number analysis uncovers a new HSCR gene: NRG3. PLoS Genet.8(5), e1002687 (2012).
- Heanue TA , PachnisV. Enteric nervous system development and Hirschsprung’s disease: advances in genetic and stem cell studies. Nat. Rev. Neurosci.8(6), 466–479 (2007).
- Emison ES , MccallionAS , KashukCSet al. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature434(7035), 857–863 (2005).
- Badner JA , SieberWK , GarverKL , ChakravartiA. A genetic study of Hirschsprung disease. Am. J. Hum. Genet.46(3), 568–580 (1990).
- Alves MM , SribudianiY , BrouwerRWet al. Contribution of rare and common variants determine complex diseases-Hirschsprung disease as a model. Dev. Biol.382(1), 320–329 (2013).
- Lake JI , HeuckerothRO. Enteric nervous system development: migration, differentiation, and disease. Am. J. Physiol. Gastrointest. Liver Physiol.305(1), G1–24 (2013).
- Jiang Q , HoYY , HaoL , NicholsBerrios C , ChakravartiA. Copy number variants in candidate genes are genetic modifiers of Hirschsprung disease. PLoS ONE6(6), e21219 (2011).
- Fitze G , SchierzM , KuhlischEet al. Novel intronic polymorphisms in the RET proto-oncogene and their association with Hirschsprung disease. Hum. Mutat.22(2), 177 (2003).
- Gui H , TangWK , SoMTet al. RET and NRG1 interplay in Hirschsprung disease. Hum. Genet.132(5), 591–600 (2013).
- Tang W , LiH , TangJet al. Specific serum microRNA profile in the molecular diagnosis of Hirschsprung’s disease. J. Cell. Mol. Med.18(8), 1580–1587 (2014).
- Revel A , AchacheH , StevensJ , SmithY , ReichR. MicroRNAs are associated with human embryo implantation defects. Hum. Reprod.26(10), 2830–2840 (2011).
- Kayano M , HigakiS , SatohJIet al. Plasma microRNA biomarker detection for mild cognitive impairment using differential correlation analysis. Biomark. Res.4, 22 (2016).
- Bong IPN , NgCC , BaharuddinP , ZakariaZ. MicroRNA expression patterns and target prediction in multiple myeloma development and malignancy. Genes Genomics39(5), 533–540 (2017).
- Gao Y , HeY , DingJet al. An insertion/deletion polymorphism at miRNA-122-binding site in the interleukin-1α 3′ untranslated region confers risk for hepatocellular carcinoma. Carcinogenesis30(12), 2064–2069 (2009).
- Dumortier O , HinaultC , Van ObberghenE. MicroRNAs and metabolism crosstalk in energy homeostasis. Cell Metab.18(3), 312–324 (2013).
- Runtsch MC , RoundJL , O’connellRM. MicroRNAs and the regulation of intestinal homeostasis. Front. Genet.5, 347 (2014).
- Sharan A , ZhuH , XieHet al. Down-regulation of miR-206 is associated with Hirschsprung disease and suppresses cell migration and proliferation in cell models. Sci. Rep.5, 9302 (2015).
- Zhu D , XieH , LiHet al. Nidogen-1 is a common target of microRNAs miR-192/215 in the pathogenesis of Hirschsprung’s disease. J. Neurochem.134(1), 39–46 (2015).
- Chen G , DuC , ShenZet al. microRNA-939 inhibits cell proliferation via targeting LRSAM1 in Hirschsprung’s disease. Aging9(12), 2471–2479 (2017).
- Li H , LiB , ZhuDet al. Downregulation of lncRNA MEG3 and miR-770-5p inhibit cell migration and proliferation in Hirschsprung’s disease. Oncotarget8(41), 69722–69730 (2017).
- Wu L , YuanW , ChenJet al. Increased miR-214 expression suppresses cell migration and proliferation in Hirschsprung disease by interacting with PLAGL2. Pediatr. Res.86(4), 460–470 (2019).
- Onkes W , FredrikR , MicciFet al. Breakpoint characterization of the der(19)t(11;19)(q13;p13) in the ovarian cancer cell line SKOV-3. Genes Chromosomes Cancer52(5), 512–522 (2013).
- Li CF , LiYC , JinJP , YanZK , LiDD. miR-938 promotes colorectal cancer cell proliferation via targeting tumor suppressor PHLPP2. Eur. J. Pharmacol.807, 168–173 (2017).
- Mi X , HeH , DengYet al. Lack of an association of miR-938 SNP in IDDM10 with human Type 1 diabetes. Diabetol. Metab. Syndr.3(1), 27 (2011).
- Cho SH , AhnEH , AnHJet al. Association of miR-938G >A polymorphisms with primary ovarian insufficiency (POI)-related gene expression. Int. J. Mol. Sci.18(6), 1255 (2017).
- Wu Y , JiaZ , CaoDet al. Predictive value of miR-219-1, miR-938, miR-34b/c, and miR-218 polymorphisms for gastric cancer susceptibility and prognosis. Dis. Markers.2017, 4731891 (2017).
- Arisawa T , TaharaT , ShiroedaHet al. Genetic polymorphisms of IL17A and pri-microRNA-938, targeting IL17A 3′-UTR, influence susceptibility to gastric cancer. Hum. Immunol.73(7), 747–752 (2012).
- Lee HA , AhnEH , JangHGet al. Association between miR-605A>G, miR-608G>C, miR-631I>D, miR-938C>T, and miR-1302-3C>T polymorphisms and risk of recurrent implantation failure. Reprod. Sci.26(4), 469–475 (2019).
- Narayanan SK , SoundappanSS , KwanEet al. Aganglionosis with the absence of hypertrophied nerve fibres predicts disease proximal to rectosigmoid colon. Pediatr. Surg. Int.32(3), 221–226 (2016).
- He J , QiuLX , WangMYet al. Polymorphisms in the XPG gene and risk of gastric cancer in Chinese populations. Hum. Genet.131(7), 1235–1244 (2012).
- He J , WangF , ZhuJet al. Association of potentially functional variants in the XPG gene with neuroblastoma risk in a Chinese population. J. Cell. Mol. Med.20(8), 1481–1490 (2016).
- He J , YangT , ZhangRet al. Potentially functional polymorphisms in the LIN28B gene contribute to neuroblastoma susceptibility in Chinese children. J. Cell. Mol. Med.20(8), 1534–1541 (2016).
- He J , WangF , ZhuJet al. The TP53 gene rs1042522 C>G polymorphism and neuroblastoma risk in Chinese children. Aging9(3), 852–859 (2017).
- Consortium GT . The genotype-tissue expression (GTEx) project. Nat. Genet.45(6), 580–585 (2013).
- Butz H , LikoI , CzirjakSet al. MicroRNA profile indicates downregulation of the TGF βpathway in sporadic non-functioning pituitary adenomas. Pituitary.14(2), 112–124 (2011).
- Li Z , JiaJ , GouJet al. Mmu-miR-126a-3p plays a role in murine embryo implantation by regulating Itga11. Reprod. Biomed. Online31(3), 384–393 (2015).
- Li H , TangJ , LeiHet al. Decreased MiR-200a/141 suppress cell migration and proliferation by targeting PTEN in Hirschsprung’s disease. Cell. Physiol. Biochem.34(2), 543–553 (2014).
- Kong YW , Ferland-MccolloughD , JacksonTJ , BushellM. MicroRNAs in cancer management. Lancet Oncol.13(6), e249–e258 (2012).
- Song FJ , ChenKX. Single-nucleotide polymorphisms among microRNA: big effects on cancer. Chin. J. Cancer30(6), 381–391 (2011).
- Hu Z , ChenJ , TianTet al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J. Clin. Invest.118(7), 2600–2608 (2008).
- Ryan BM , RoblesAI , HarrisCC. Genetic variation in microRNA networks: the implications for cancer research. Nat. Rev. Cancer10(6), 389–402 (2010).
- Iwai N , NarabaH. Polymorphisms in human pre-miRNAs. Biochem. Biophys. Res. Commun.331(4), 1439–1444 (2005).
- Xu J , HuZ , XuZet al. Functional variant in microRNA-196a2 contributes to the susceptibility of congenital heart disease in a Chinese population. Hum. Mutat.30(8), 1231–1236 (2009).
- He J , ZouY , LiuXet al. Association of common genetic variants in pre-microRNAs and neuroblastoma susceptibility: a two-center study in Chinese children. Mol. Ther. Nucleic Acids11, 1–8 (2018).
- Iwata J , HosokawaR , Sanchez-LaraPAet al. Transforming growth factor-beta regulates basal transcriptional regulatory machinery to control cell proliferation and differentiation in cranial neural crest-derived osteoprogenitor cells. J. Biol. Chem.285(7), 4975–4982 (2010).
- Zhang M , CaoSR , ZhangR , JinJL , ZhuYF. The inhibitory effect of salvianolic acid B on TGF-beta1-induced proliferation and differentiation in lung fibroblasts. Exp. Lung Res.40(4), 172–185 (2014).
- Qian T , ShiS , XieL , ZhuY. miR-938 promotes cell proliferation by regulating RBM5 in lung adenocarcinoma cells. Cell. Biol. Int.44(1), 295–305 (2020).