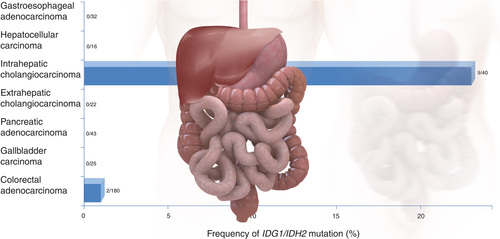
Recent research has revealed an association between IDH1 mutation and a subgroup of incurable cholangiocarcinomas. It is hoped that the discovery could not only provide a new target for novel drug development, but also influence the development of a novel biomarker test for the disease.
In a recent study carried out by researchers at the Massachusetts General Hospital Cancer Center (MA, USA), IDH1 and IDH2 mutations were found to be associated with bile duct cancers of intrahepatic origin. ‘The high prevalence of IDH1 mutations suggests a new target for therapeutic intervention in a significant number of patients with inoperable intrahepatic bile duct tumors,’ explained lead author Darrell Borger when speaking to Personalized Medicine. Borger is an Instructor of Medicine at Massachusetts General Hospital. The team also discovered that IDH1 mutation is associated with elevated intratumoral levels of 2-hydroxyglutarate, which could be a potential biomarker for this disease. The study was published online ahead of print in December 2011 in the journal The Oncologist.
Bile duct cancer is diagnosed in approximately 12,000 patients per year in the USA, with 90% of cases detected at advanced stage with poor prognosis. Average patient survival time is approximately less than 1 year owing to the impossibility of surgical intervention and the limited benefits of chemo- and radio-therapy. Understanding the molecular basis of this disease is crucial to opening up new diagnostic and treatment possibilities.
In this recent study, a total of 287 tumors from patients with gastrointestinal cancers were genotyped during routine clinical evaluation. ‘We have developed a genotyping platform that coamplifies regions of essential cancer genes that harbor known site-specific mutations,’ elaborated Borger. ‘The hotspot mutations of interest are interrogated through a one-base extension step, allowing us to simultaneously evaluate over 120 site-specific mutations across more than 14 cancer genes.’ Borger added that by focusing on the most therapeutically relevant genes, his team have been able to broadly apply this platform across a range of gastrointestinal malignancies, such as in this recent study.
While IDH1 mutations were found to be present in only 2% of the common gastrointestinal tumors included in this study, the abnormality was found in 25% of the rarer malignancy, bile duct carcinoma. This discovery led the researchers to genotype an additional 75 gallbladder and bile duct tumors; IDH1 and IDH2 mutations were detected in 23% of intrahepatic bile duct tumors. The mutations were absent from the extrahepatic bile duct and gallbladder tumors.
A subsequent analysis of frozen tissue specimens revealed that IDH1 mutation was associated with dramatically elevated intratumoral levels of 2-hydroxyglutarate. ‘It will be of interest to determine whether blood levels of 2-hydroxyglutarate can serve as a biomarker for IDH gene status as well as a means of following treatment effects when targeted agents become available,’ explained Borger.
However, investigating a rare tumor type, such as cholangiocarcinoma, presents some challenges, as Borger told Personalized Medicine: ‘One of our biggest challenges is access to patient tumor samples of sufficient quality for detailed molecular analyses, or of sufficient quantity that provides the power to correlate a molecular signature with clinical outcome.’ He concluded that, in order for insights into cancer genetics to impact on personalized medicine, developments in bioinformatic approaches are required: ‘Being able to generate and interpret this information in a timely manner so that it can be used by the clinicians to direct clinical care is currently one of our biggest challenges.’
– Written by Sarah Miller
Sources: Massachusetts General Hospital news release: www.massgeneral.org/about/pressrelease.aspx?id=1430; Borger DR, Tanabe KK, Fan KC et al. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist 17(1), 72–79 (2012).
A biomarker-based test for diagnosing facioscapulohumeral muscular dystrophy (FSHD) has moved one step closer owing to the latest in a series of discoveries by a group of scientists from the Fred Hutchinson Cancer Research Center (Seattle, WA, USA).
The group have already identified key genes and proteins in addition to several discoveries regarding the disease mechanisms of FSHD; the third most common type of inherited muscular dystrophy.
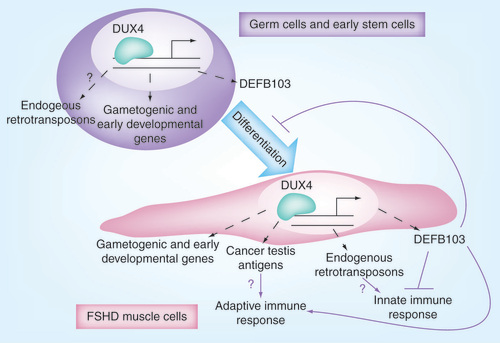
In the most recent study, evidence has been accumulated to support how the transcription factor DUX4 can cause FSHD. Genes associated with germline and early stem-cell development were established as targets of DUX4, a leading candidate gene for FSHD. Genes that are regulated by DUX4 can be consistently detected in FSHD-affected muscle but are absent from controls, suggesting that abberant expression of DUX4 is a causal factor for this type of muscular dystrophy. details the mechanism of action of DUX4.
DUX4 target genes are expressed in FSHD skeletal muscle. DUX4 targets are candidate biomarkers for FSHD and suggest disease mechanisms.
FSHD: Facioscapulohumeral muscular dystrophy.
Reproduced with permission from source 1 (Geng LN et al.)
Whether DUX4 is the sole cause of FSHD remains to be proven. However, one of the investigators, Stephen Tapscott explained; ‘This study is a significant step forward by solidifying that the DUX4 transcription factor causes this disease, while offering a number of viable mechanisms for why the muscle is damaged.’
It is hoped that an antibody- or RNA-based test could be developed for the diagnosis of FSHD from a muscle tissue biopsy. In addition, these tests could be used to determine the efficacy of treatments for FSHD.
– Written by Emily Reeve
Sources: Geng LN, Yao Z, Snider L et al. DUX4 Activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell 22(1), 38–51 (2012); Fred Hutchinson Cancer Research Center news release: www.fhcrc.org/content/public/en/news/releases/2012/01/genes-diseaase-muscular-dystrophy.html
A recent study carried out by the Moffitt Cancer Center (FL, USA) has revealed that a malignancy-risk gene signature, previously applied to predict breast cancer risk, may also have predictive and prognostic value in early-stage non-small-cell lung cancer (NSCLC). The gene signature could be used to identify NSCLC patients most at risk of disease relapse, enabling more effective targeting of adjuvant chemotherapy to individuals most likely to benefit. The findings were published in late December 2011, in the Journal of the National Cancer Institute.
NSCLC accounts for approximately 80–90% of all lung cancer cases, and patients have a 30–50% chance of relapse following surgery. While adjuvant chemotherapy has increased survival rates, not all patients benefit. The 5-year survival rate ranges from 40 to 70%.
In this recent study, three NSCLC microarray datasets were analyzed (n = 692) in order to assess the ability of the malignancy-risk gene signature to predict overall survival (OS) in NSCLC patients. Researchers employed principal component analysis to determine an overall malignancy-risk score, and an interaction model was used to investigate a statistically significant interaction between the gene signature and adjuvant chemotherapy.
The malignancy-risk gene signature was significantly associated with OS (p < 0.001) in NSCLC patients. The prognostic value of the gene signature was demonstrated by the observation that low malignancy-risk score patients who did not receive adjuvant chemotherapy had increased OS compared with those with a high malignancy-risk score. High malignancy-risk score patients who received adjuvant chemotherapy had their OS significantly increased (p = 0.03). The team also reported a statistically significant interaction between the gene signature and adjuvant chemotherapy (p = 0.02).
It is hoped that the malignancy-risk gene signature investigated in this recent study could be used to identify early-stage NSCLC patients with a predicted low OS who could then be recommended for adjuvant chemotherapy. A future, larger study is planned using patient-donated tissue samples.
– Written by Sarah Miller
Source: Chen DT, Hsu YL, Fulp WJ et al. Prognostic and predictive value of a malignancy-risk gene signature in early-stage non-small cell lung cancer. J. Natl Cancer Inst. 103(24), 1859–1870 (2011).
A 5-year multicenter prospective study performed at Cleveland Clinic (OH, USA) has provided new insights into the genetics of Cowden syndrome (CS) and thyroid cancer through studies of patients with CS or CS-like disease. Germline PTEN, SDHx and KLLN alterations were found to increase thyroid cancer risk. Researchers hope that the findings could eventually lead to extended genetic testing for children with thyroid cancer in order to personalize therapy using gene-specific treatments.
Germline mutations in PTEN and SDHx, in addition to KLLN epimutation, are known to cause CS and CS-like disease, of which thyroid cancer is thought to be a key component. Amongst other findings, this recent study demonstrated that individuals with CS are at a 70-fold higher risk of thyroid cancer than the general population. ‘This highlights the need to continue to accrue data to determine if current surveillance guidelines for thyroid cancer in CS patients are adequate; for example, at present it has no provisions for pediatric patients,’ commented lead author Charis Eng when speaking to Personalized Medicine. Eng is the chair and founding director of the Genomic Medicine Institute of Cleveland Clinic’s Lerner Research Institute (OH, USA).
This recent study analyzed 2723 CS and CS-like patients. All participants underwent PTEN genotyping. Those without PTEN mutations were analyzed for SDHx mutations, and those without SDHx alterations were analyzed for KLLN epimutation. ‘By systematically studying patients who were PTEN-mutation negative, we have found two other genes associated with CS and from that, the discovery that all three result in increased thyroid cancer risk, which forms the basis for subsequent research,’ explained Eng.
In addition to confirming a genetic link between CS and thyroid cancer, the Cleveland Clinic researchers found that different genetic alterations were associated with different clinical histological subtypes of thyroid cancer. PTEN mutation-positive CS and CS-like patients were found to have an increased risk of follicular thyroid cancer, while those harboring SDHx or KLLN alterations were at an increased risk of papillary thyroid cancer.
These three genes are not only associated with increased thyroid cancer risk; for example, PTEN mutation also increases a patient’s risk of breast, renal, endometrial and colon cancers. Eng suggests that upon discovery of PTEN alterations in a patient, increased screening for the gene-specific range of cancers should be commenced.
In addition, PTEN, SDHx and KLLN belong to different cell pathways, potentially allowing different targeted treatments to be used depending on the genetic alteration involved.
Eng explained to Personalized Medicine that the collection of good quality clinical phenotypic data was made more challenging due to CS being an uncommon disease, often overlooked by nonspecialists: ‘One has to ensure that we are comparing apples to apples (i.e., that we enrolled patients who indeed had CS clinical features). I am fortunate to have a dedicated and meticulous team who works closely with our recruiting sites to ensure that every patient’s medical records are reviewed and features documented only when confirmed by pathology reports or clinical notes,’ concluded Eng.
– Written by Sarah Miller
Sources: Cleveland clinic news release: http://my.clevelandclinic.org/media_relations/library/2011/2011-12-23-cleveland-clinic-researcher-discovers-genetic-cause-of-thyroid-cancer.aspx; Ngeow J, Mester J, Rybicki LA, Ni Y, Milas M, Eng C. Incidence and clinical characteristics of thyroid cancer in prospective series of individuals with Cowden and Cowden-like syndrome characterized by germline PTEN, SDH, or KLLN alterations. J. Clin. Endocrinol. Metab. 96(12), E2063–E2071 (2011).