Abstract
An important part of teaching students how to use the BLAST tool for searching large sequence databases, is to train the students to think critically about the quality of the sequence hits found — both in terms of the statistical significance and how informative the individual hits are. This paper describes how generating truly random sequences by throwing dice can be used to illustrate how unrelated sequences may be found by BLAST, how to judge the statistical significance of the hits, and how the database size influences the statistics.
Introduction
Searching large sequence databases such as GenBank (CitationBenson, Karsch-Mizrachi et al, 2007) and CitationUniProt (UniProt_Consortium 2007) is a core element within the field of bioinformatics, as well as in the more general field of molecular biology. When teaching students (from first year students to PhD level) how to use the BLAST tool to perform sequence based searches, it is of vital importance that the students develop a critical sense of how to evaluate the significance of the results. This is not trivial; quite often, one obtains results that on the surface look convincing, but will lead to wrong conclusions if the statistics are ignored. This paper describes how the use of polyhedral dice in a teaching exercise illuminates this issue, by having the students search the databases using truly random sequences as search queries. These are generated by throwing four-sided dice for DNA sequences and twenty-sided dice for Protein-sequences.
One of the most important skills for both bioinformaticians and wet-lab biologists working with sequence data is to be able to infer the function of an unknown DNA or protein sequence. This is typically done by searching a large database of known sequences. The input sequence is compared to each sequence in the database using an algorithm known as local pair-wise alignment (CitationSmith and Waterman 1981). The standard tool for doing this is BLAST (Basic Local Alignment Search Tool; (CitationAltschul, Gish et al, 1990; Altschul, Madden et al, 1997)). The main reason BLAST is the tool of choice is its built-in heuristics that can quickly eliminate the sequences that are not likely to produce significant result, thus reducing the search space and offering very fast search-times.
Teaching BLAST
One of the main issues when teaching students the use of BLAST is how to interpret the significance of the results. The key problem is that if the database is large enough (e.g. billions of DNA “letters”) there is a inherent probability of picking up a short, perfectly matching -yet unrelated- hit to any given input sequence. In order to address this issue the BLAST algorithm calculates a so-called “expect-value” (e-value). This number estimates how many unrelated (random) hits of equal or better quality the users should expect given the database size.
The problem with using the e-values in a teaching setting is that it is very “black-box” like and that the students can be fooled by the apparent good quality of a sequence match when looking at the sequence alignment itself (see compared to ).
Figure 1 Examples of insignificant BLAST hit. Query” = input sequence (fragment). “Sbject” = matching sequence found in the database (fragment). E-values (“expect”) highlighted in red.
Both the DNA and protein query sequences are randomly generated, using the approach described in this paper.
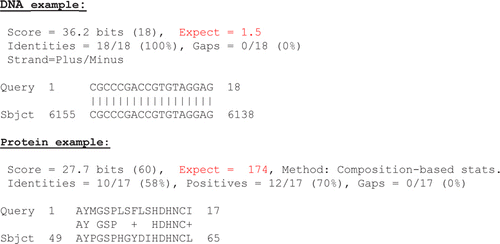
I have addressed this issue by having my students investigate the kind of results that can be obtained when using truly random sequences as input data. The rationale behind this is that it is - in my experience - truly an eye-opener that BLAST can produce results that may look convincing at the first glance, but which are not biological meaningful at all.
As the first part of the exercise the students construct short sequence fragment (DNA or protein) by throwing four- or twenty-sided dice (see ). Since the students have created the sequences themselves, it should be evident to them that the results found cannot be “true hits” (that is, a hit from an evolutionary related sequence). When they go through the list of hits and compare the quality of the hits at the sequence level (see ) to the e-value, the importance of evaluating the statistical significance of the result becomes self-evident. The random sequences are also good for highlighting the issue of how the e-value relates to the database size. The students BLAST the same sequences against both the Human database (genome + mRNA; ∼5 gigabases) and “NR” database (∼20 gigabases) at NCBI. Since the “NR” database also contains the human sequences, the hits found in the human database, will also be found here, but now the e-value will be four times higher (since the e-value for a truly random hit doubles each time the database size double). In combination with showing a few extreme-value plots in the lecture prior to the exercise, this nicely sums up how the e-value of a hit depends on the database.
The DNA example in figure 2 is the Yeast HTA2 gene matched against the HTA2 homolog from Kluyveromyces lactis. The protein example is an unknown protease from an uncultivable soil microorganism (sequenced PCR fragment) matched against a Bacteroides S8 protease.
Figure 2 Examples of significant BLAST hit. “Query” = input sequence (fragment). “Sbject” = matching sequence found in the database (fragment). E-value (“expect”) highlighted in red.

Once the students have explored the issue of random unrelated hits, the students move onto BLASTing real biological sequences (see ). By doing this after working with the random sequences, it will quickly become clear that a true biological hit differs greatly from the random hits in the following:
The alignments are much longer and the e-value is much lower (long alignment ⇒ high alignment score ⇒ low e-value; see ).
Table 1 Generation of truly random DNA/protein sequences. Please observe that nucleotide and amino-acid frequencies produced by throwing dice do not reflect the frequency distribution from true biological sequences. This is especially true for protein sequence, where the background frequencies are very skewed (for eukaryotic DNA the 25% per base frequency is a decent approximation). However, I have chosen to deliberately ignore this issue for the BLAST exercise described here — in order to focus only on the issue of randomness and significance, and avoid “information overload”. I deal with the issue of amino-acids frequencies in great details later in my “Introduction to Bioinformatics” course, when we go into matrix methods, pseudo-count correction and Logo plots.
In the case of BLASTing a sequence of known function, the high-scoring hits will agree on that function. (Or at least not disagree; BLASTing a Yeast gene like HTA2 against the NR database will produce two kind of perfect match hits: short well described GenBank entries, and hits against large chromosomal fragments from Yeast with no individual genes assigned).
Figure 3 How the length of the alignment affect the e-value. The two examples shown here are both subsets of the protein sequence (a protease) from . As the length of the input sequence decreases, the significance of the result also decreases.

“Informative hits”: In many cases BLAST will return a lot of very significant hits that are all “gene of unknown function” or “hypothetical protein”. This is a good opportunity to have a discussion with the students of what is an “informative hit” and what is not. (On a side note, I should mention that from day one in my Introduction to Bioinformatics course I put a large effort into teaching my students about data quality — for example how to judge if a GenBank or UniProt entry is reliable or not).
DNA vs. Protein: I have a small collection of protease sequences (sequenced directly from un-cultivatable soil micro-organism) that are not present in any public database. It is impossible to find any significant hits at the DNA level, whereas translating the sequences and BLASTing at the protein level will quickly reveal the protein function. This also offers a good opportunity to discuss the differences in the heuristics of BLASTN and BLASTP which are profound but which appears to be unknown to many people using BLAST (even including research collaborators).
It will also be easy to see that for DNA sequences the e-value distribution is bi-modal: usually some very significant hit (e.g. < 1e-30, using the software notation) and a lot of short fragments with insignificant e-value (e.g in the 1e-3 – 1e-10 range) that resembles the random sequence hits. For protein sequences it will become apparent that the e-value distribution is much more smooth with a lot of intermediate e-values. This can use used as a starting point of the important discussion of what is a significant e-value? (Rule of thumb of protein sequences: e-value must be lower than 1e-5). For a further discussion of the issue of alignment at the DNA level versus the protein level please see (CitationWernersson and Pedersen 2003).
In conclusion, it is my impression that having the students starting by using random sequences enhances their ability to judge the significance of a BLAST hit, and provides a good starting point for moving onto the analysis of real biological sequences. Furthermore I believe that having the students throwing dice instead of using a computer program to generate random sequences enhances their trust in the randomness of the sequences and makes the eye-opening experience even bigger. Using “exotic” polyhedral diceFootnote1 (known from role-playing games, such as “Dungeons and Dragons”), further makes the computer exercise something the students remember.
An English language version of the computer exercise described here is in preparation and will be available from the authors homepage: http://www.cbs.dtu.dk/~raz/
Notes
1 These kinds of dice is available a virtually all shops with role-playing books and equipment. In Denmark I can recommend the “Fantask” shop in central Copenhagen.
References
- AltschulS. F., W.Gish et al. (1990) Basic local alignment search tool. J Mol Biol 215(3) 403-410
- AltschulS. F., T. L.Madden et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17) 3389-3402
- BensonD. A., I.Karsch-Mizrachi et al. (2007). GenBank. Nucleic Acids Res 35(Database issue): D21-5
- SmithT. F. and M. S.Waterman (1981) Identification of common molecular subsequences. J Mol Biol 147(1) 195-7
- UniProt_Consortium (2007) The Universal Protein Resource (UniProt). Nucleic Acids Res 35(Database issue) D193-7
- WernerssonR. and A. G.Pedersen (2003) RevTrans: Multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res 31(13) 3537-3539