Abstract
Purpose: The purpose of this study was to identify reference genes showing stable expression in chondrocytes cultured under several different thermal environments and in different culture systems.
Materials and methods: Human articular chondrocytes were cultured by monolayer or pellet culture system at 32 °C, 37 °C, and 41 °C for 3 days. Thereafter, the total RNA was extracted, and quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) was performed. The qRT-PCR data was analysed using three different algorithms (geNorm, NormFinder, and BestKeeper) to identify reference genes exhibiting stable expression from among the seven candidate reference genes (B2M, ACTB, GAPDH, HSPCB, RPL13a, YWHAZ, and 18S).
Results: The candidate reference genes, except for HSPCB and YWHAZ, showed systematic variations in expression. In the monolayer culture, RPL13a was the most stable gene identified using NormFinder and BestKeeper; on using geNorm, ACTB and GAPDH showed the highest expression stability. In the pellet culture, ACTB was the most stable gene identified using NormFinder and BestKeeper, whereas GAPDH and RPL13a were the most stable reference genes as determined using geNorm. In the combined group, B2M and GAPDH were the most stable genes identified using geNorm, whereas RPL13a and YWHAZ were the most stable as per NormFinder and BestKeeper, respectively. The best combination of two candidate reference genes among all the groups determined using NormFinder was RPL13a and YWHAZ.
Conclusion: The combination of RPL13a and YWHAZ might be suitable as reference genes for human chondrocytes cultured at 32 °C, 37 °C, and 41 °C in monolayer, pellet, or combined cultures.
Introduction
Articular cartilage, which exists at the epiphysis of articular joints, is composed of an extracellular matrix (ECM) with sparse distribution of highly specialised cells called chondrocytes. Since the chondrocytes are surrounded by the ECM and exposed to low oxygen conditions in the body, chondrocytes are often grown in three-dimensional (3D) cultures in a low-oxygen environment to mimic in vivo conditions [Citation1]. Moreover, chondrocytes reside at a relatively low basal temperature (approximately 32 °C) in the knee joint [Citation2,Citation3]. However, few studies have considered the effect of in vivo temperature in in vitro models. Recently, we reported that at a culture temperature of 32 °C, chondrocytes produce an equivalent amount of ECM to that produced at 37 °C and that the ECM production was inhibited at 41 °C [Citation4]. Serrat et al. [Citation5] reported that culturing chondrocytes at 39 °C promotes proliferation and ECM synthesis. However, the effects of the culture temperature on chondrocyte metabolism remain unclear.
Thermal stimuli (e.g. hyperthermia) has been used in the treatment of diseases, including malignancies [Citation6] and osteoarthritis [Citation7]. Mild thermal stimulation induces the expression of heat shock proteins (HSPs) and increases tolerance to stress [Citation8–10]. Therefore, the identification of the effects of thermal stimuli on chondrocyte metabolism opens new avenues for the treatment for several diseases.
Quantitative real-time polymerase chain reaction (qPCR) is a powerful and sensitive method for analysing mRNA expression. An important step in the analysis of gene expression using this method is the normalisation of the data to compensate for differences in the purity and concentration of RNA among samples [Citation11,Citation12]. In qPCR, this is generally performed by normalising the expression of the gene of interest against that of an endogenous reference gene, whose expression is stable across tissues, cell types, and conditions. However, it is likely that a gene that fulfils all of these criteria does not exist [Citation13]. Therefore, it is necessary to identify an appropriate reference gene for each experimental condition.
A recent study identified and validated reference genes suitable for the analysis of gene expression in chondrocytes cultured under hypoxic conditions [Citation14]. However, to the best of our knowledge, there have been no studies investigating the effects of temperature on the reference genes in human chondrocytes. Here we hypothesised that variations in temperature would affect the expression levels of genes whose expression is considered as stable in human chondrocyte cultures. The purpose of this study was to identify reference genes whose expression is stable in chondrocytes cultured at various different temperatures and in different culture systems.
Materials and methods
Chondrocyte isolation
Human articular cartilage was obtained from a femoral head extracted while performing a bipolar hip arthroplasty on a 62-year-old woman. Chondrocytes were isolated by a previously described method [Citation15]. The Ethics Committee of the Faculty of Medicine, Kyoto University approved the procedure, and informed consent was obtained from the donor. The isolated cells were resuspended in fresh Dulbecco’s modified Eagle medium/Ham’s F12 (DMEM/HamF12) (Nacalai Tesque, Kyoto, Japan) containing 10% fetal bovine serum (FBS) (Hyclone, Logan, UT), 50 U/mL penicillin (Nacalai Tesque), and 50 μg/mL streptomycin (Nacalai Tesque), and were seeded in tissue culture dishes. The cells were expanded in a CO2 incubator (5% CO2; 37 °C; and 95% humidity) until passage two. The cells were divided into monolayer and pellet culture groups, each of which was subdivided into three subgroups based on the incubation temperature (32 °C, 37 °C, and 41 °C).
Monolayer culture
The expanded chondrocytes were subcultured at 1.5 × 104 cells/cm2, and then pre-cultured at 37 °C for three days in a CO2 incubator. After pre-culture, the cells were transferred into separate CO2 incubators set at 32 °C, 37 °C, and 41 °C, respectively, and were incubated for 3 days. The temperatures used were designated based on the normal intra-articular temperature (32 °C) [Citation2,Citation3], internal body temperature (37 °C), and threshold temperature for mammalian cell survival (41 °C) [Citation16,Citation17].
Pellet culture
To provide a 3D environment, a pellet culture system was used in this study. Pellet culture is a simple and widely used technique to evaluate chondrogenic potential and to study the signalling pathways involved in chondrogenesis [Citation18,Citation19]. The expanded chondrocytes were trypsinised and then washed with DMEM/HamF12, re-suspended in chondrogenic medium (chondrogenic basal medium plus ITS + supplement, ascorbate, dexamethasone, l-glutamine, sodium pyruvate, proline, and GA-1000 from Lonza, Walkersville, MD) supplemented with 10 ng/mL of recombinant human transforming growth factor-beta 3 (R&D Systems, Minneapolis, MN). Aliquots of 2.5 × 105 cells in 500 μL of chondrogenic medium were centrifuged at 250 × g for 5 min in 15-mL polypropylene conical tubes. The pelleted cells were pre-cultured at 37 °C in a CO2 incubator for 3 days, and the pellets were cultured at 32 °C, 37 °C, and 41 °C for 3 days.
Total RNA extraction
The cells in the monolayer cultures (n = 8 culture dishes/subgroup) and the chondrocyte pellets in the pellet cultures (n = 8 pellets/subgroup) were harvested after 3 days. Total RNA was extracted using the RNeasy Mini Kit (Qiagen, Valencia, CA), as per the manufacturer’s protocol, and were purified by performing RNase-free DNase on-column incubation. Total RNA purity was confirmed by measuring the A260/A280 ratio (>2.0) for all the samples by using the NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, MA). The RNA was stored at −80 °C until further use.
Real-time qRT-PCR
RT-PCR was performed using the ReverTra Ace qPCR RT Kit (Toyobo, Osaka, Japan), according to the manufacturer’s protocol. Total RNA (350 ng) was reverse-transcribed for 15 min at 37 °C to synthesise cDNA, followed by incubation at 98 °C for 5 min to deactivate the enzymes. The RT-PCR products were stored at −30 °C.
qPCR was performed using the Applied Biosystems 7500 Real-Time PCR System (Life Technologies, Carlsbad, CA). cDNA templates corresponding to 4.375 ng of total RNA were amplified using Power SYBR Green PCR Master Mix (Life Technologies) in 25 μL reactions containing 1 × Power SYBR Green PCR Master Mix, 0.2 μM of each gene-specific primer (), and deionised water. The mixture was initially heated to 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 15 s, and annealing and extension at 60 °C for 60 s. Following amplification, melt curve protocols were performed to ensure that primer-dimers or non-specific products, if any, were eliminated or minimised. The candidate reference genes evaluated in this study are listed in . Selection of the genes belonging to different functional and abundance classes was performed carefully, which significantly reduced the risk of genes being co-regulated. The primers used are described in Andersen et al. [Citation20], Studer et al. [Citation21], and Curtis et al. [Citation22], and are listed in . PCR efficiency for each primer pair was determined ().
Table I. Primer sequences for candidate reference genes.
Table II. Candidate reference genes evaluated in this study.
Data analysis
All the corresponding qRT-PCR data were analysed using the 7500 System Sequence Detection Software version 2.0.4 (Life Technologies). The quantification cycle (Cq) is defined as the number of cycles needed for the fluorescence to reach a specific threshold level of detection, and is inversely correlated with the amount of template nucleic acid present in the reaction [Citation23]. Data analysis was performed using the geNorm [Citation13], NormFinder [Citation20], and BestKeeper [Citation24] software packages. geNorm is a Visual Basic Application for Microsoft Excel that ranks the reference genes on the basis of stepwise elimination of the least stable gene. Expression stability (M value) is defined as the average pairwise variation of a given gene with all the other candidate reference genes. NormFinder, an add-in for Microsoft Excel, ranks the reference genes using a model-based approach, and identifies the best candidate gene based on minimal combined inter- and intragroup expression variations. BestKeeper, an Excel-based tool, ranks reference genes on the basis of repeated pairwise correlation and regression analysis of a given gene with all the other tested candidate reference genes. The genes with the lowest stability values have the most stable expression in these software programs. The expression stabilities of several candidate reference genes in human chondrocytes cultured in different thermal environments were evaluated using these three software programs.
Results
RNA transcription levels of candidate reference genes in various temperature environments
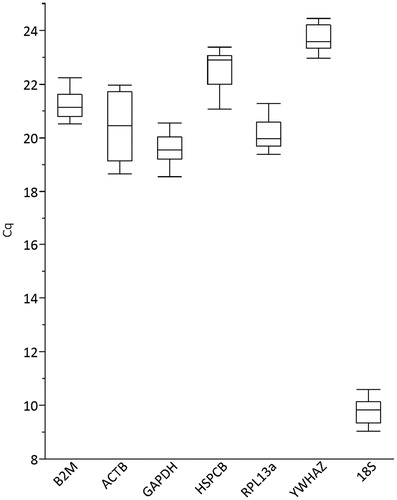
The overall RNA transcription pattern of seven candidate reference genes in human chondrocytes grown in various thermal environments and culture systems is represented in . Of the genes examined, 18S ribosomal RNA (rRNA) showed the highest transcription level (low Cq value). Ribosomal RNAs including 18S rRNA have been reported to be unsuitable for normalisation, as these genes are overexpressed in comparison to other mRNAs, and because they are transcribed by RNA polymerase I [Citation25]. These characteristics make it difficult to correct for sample-to-sample variations in qRT-PCR and also affect the accuracy of the data. Therefore, 18S rRNA was excluded from further studies.
Figure 1. Overall RNA transcription patterns of candidate reference genes under different thermal environments and culture systems. The box and whisker diagrams show the quantification cycle (Cq) values for each gene. The band inside each box represents the median Cq, while the bottom and top are the first and third quartiles, respectively. The lines extending from each box (whiskers) indicate the maximum and minimum values.
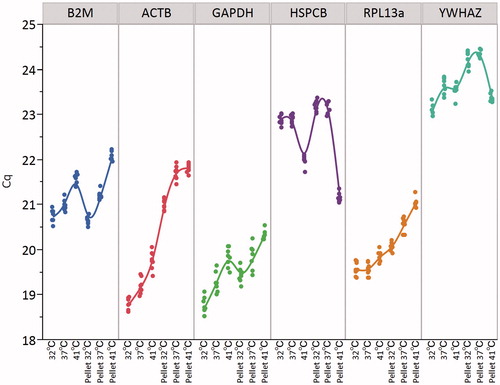
The RNA transcription profiles of the candidate reference genes in sample subgroups are represented in . As expected, the RNA transcription level of each gene varied among the subgroups, indicating that the temperature and culture system used (monolayer or pellet) affected the RNA transcription profiles of these candidate reference genes. Interestingly, with the exception of HSPCB and YWHAZ, each gene showed systematic variation, which tended to decrease the RNA transcription level (based on increased Cq value) at higher temperatures across the sample groups.
Figure 2. Effect of temperature and culture conditions on RNA transcription profiles of the candidate reference genes. The dots represent the Cq values of genes across different subgroups. Each curve is a cubic spline with a default lambda of 0.05 and standardised X-value.
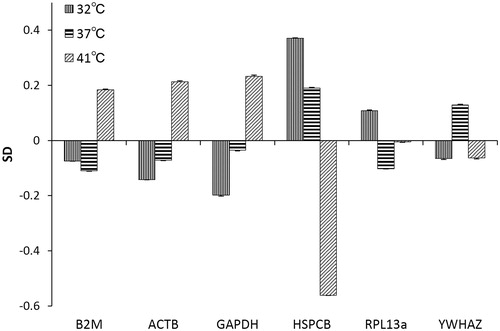
The standard deviation (SD) of each candidate reference gene among the different subgroups of the monolayer culture is shown in . The expression of ACTB and GAPDH showed similar systematic variation, and that of HSPCB showed a large variation, unlike that observed for the other genes. RPL13a and YWHAZ showed small and somewhat compensatory variation.
Ranking of the candidate reference genes
Gene expression stability was ranked using geNorm, NormFinder, and BestKeeper, which are three commonly used algorithms for selecting suitable reference genes. shows the ranking and stability values for the monolayer, pellet, and combined groups. Rankings in NormFinder and BestKeeper were very similar in all the groups, although this trend was not observed in case of geNorm. In monolayer culture, RPL13a was the most stable gene, while GAPDH and HSPCB were ranked low by the NormFinder and BestKeeper algorithms. By contrast, geNorm analysis showed that ACTB and GAPDH were the most stable genes and that HSPCB was the least stable gene. In pellet culture, ACTB was the most stable gene, and HSPCB was the least stable gene in NormFinder and BestKeeper. However, geNorm analysis showed that GAPDH and RPL13a were the most stable genes and that HSPCB was the least stable gene. In the combined group, RPL13a and YWHAZ were the most stable genes as per NormFinder and BestKeeper, respectively, while ACTB was the least stable. As per geNorm, B2M and GAPDH were the most stable genes and HSPCB was the least stable.
Table III. Stability values and ranking of candidate reference genes obtained using different normalisation softwares.
shows the best combination of two candidate reference genes based on NormFinder analysis. In all the groups, the combination of RPL13a and YWHAZ was the most stable compared to any single reference gene in the candidate genes studied.
Table IV. Two-gene combinations with the highest stability values (in parentheses) obtained using NormFinder.
Discussion
The findings of this study clearly demonstrate the effect of temperature and culture conditions on the stability of reference genes in human chondrocytes. Importantly, several candidate reference genes showed similar systematic variations in expression in response to temperature. shows the results of repeated pairwise correlation analysis of the candidate reference genes in monolayer culture by using BestKeeper. The analysis showed strong correlation among the genes, particularly between ACTB and GAPDH (r = 0.988). These results, which are indicative of systematic expression variations in the reference genes, make it difficult to calculate gene stability accurately. The underlying principle of geNorm is that it assumes that reference genes are not co-regulated, and that the expression ratio of any two reference genes is constant across samples [Citation13]. Consequently, geNorm results could be misleading when reference genes show similar responses to experimental treatments [Citation20]. In this study, the rankings obtained using geNorm might have resulted from the co-regulation of the candidate reference genes.
Table V. Repeated pair-wise correlation among reference genes in monolayer culture based on BestKeeper analysis.
The ranking of HSPCB was low in all the groups. HSPCB encodes a member of the heat shock protein 90 (Hsp90) family that aids in protein folding and quality control for a large number of client proteins. One of the causes of the low ranking for this gene might be that its expression was induced by heat shock only at 41 °C, but not at 32 °C or 37 °C. We confirmed that HSP70, which is one of the HSP members, was not induced at 32 °C or 37 °C, but that significant induction was observed at 41 °C (data not shown). Therefore, it is possible that HSPCB expression shows a similar response to temperature in chondrocytes.
Recently, a novel strategy for the identification of stably expressing reference genes for normalisation of qRT-PCR data was reported [Citation13]. However, this method is effective only if expression variance is reduced when genes are tested in combination [Citation20]. shows the inter-subgroup variation in monolayer culture analysed using NormFinder. The combination of ACTB and GAPDH showed the most stable expression as per the geNorm analysis (). Despite this, these genes might not be suitable for normalisation because they demonstrated similar systematic variation that would not be reduced if the genes were assessed in combination. On the other hand, RPL13a and YWHAZ, which was the most stable gene combination identified using NormFinder, might show reduced variance. Therefore, we considered this a suitable combination. NormFinder was able to take into account systematic differences between intergroup variations, and was less affected by the correlated expression of the candidate reference genes [Citation20].
The best combination of the two reference genes identified using NormFinder in all the groups was RPL13a and YWHAZ. RPL13a encodes a ribosomal protein that is a component of the 60S subunit in the ribosome. This gene has previously been shown to be a reliable reference gene in normoxic- and hypoxic-cultured human chondrocytes [Citation14] as well as in other cell types [Citation26]. YWHAZ is a signal transduction protein that binds to phosphorylated serine residues on a variety of signalling molecules. YWHAZ showed no systematic variation (), and its standard deviation values were relatively low (). Taken together, the combination of RPL13a and YWHAZ might be suitable as reference genes for human chondrocytes cultured at 32 °C, 37 °C, and 41 °C in monolayer, pellet, or combined cultures.
We identified three limitations in this study. First, our results were derived by analysing the cells obtained from only one patient. Therefore, our data only indicate expression stability in response to experimental treatments, and do not take into account variations among individuals, tissues, or other cell types. Second, although we evaluated seven candidate reference genes belonging to different functional classes, these genes might not be representative. Third, our evaluations were conducted under normoxic conditions, and the expression of these reference genes may well be different in cells cultured under low oxygen conditions. Chondrocytes show enhanced ECM synthesis [Citation27] and maintain their chondrogenic differentiation state [Citation28] in hypoxic culture conditions. Moreover, chondrocytes exist in a low oxygen environment in vivo [Citation29,Citation30]. Therefore, further studies on the effect of oxygen tension on reference gene stability in chondrocytes are warranted.
Conclusion
Here we evaluated suitable reference genes for human chondrocytes cultured in several different thermal environments in monolayer and pellet culture systems. Our results indicate that several candidate reference genes showed systematic variations in expression in response to temperature. We conclude that the combination of RPL13a and YWHAZ is probably suitable as reference genes for human chondrocytes cultured at 32 °C, 37 °C, and 41 °C in monolayer, pellet, or combined cultures.
Declaration of interest
This study was supported in part by a Grant-in-Aid for the Japan Society for the Promotion of Science (JSPS) Research Fellows (number 820130600018), JSPS KAKENHI Grant-in-Aid for Scientific Research (A) (number 25242055), and JSPS KAKENHI Grant-in-Aid for Challenging Exploratory Research (number 25560258).
The authors alone are responsible for the content and writing of the paper.
References
- Foldager CB, Nielsen AB, Munir S, Ulrich-Vinther M, Soballe K, Bunger C, et al. Combined 3D and hypoxic culture improves cartilage-specific gene expression in human chondrocytes. Acta Orthop 2011;82:234–40
- Oosterveld FG, Rasker JJ. Treating arthritis with locally applied heat or cold. Semin Arthritis Rheum 1994;24:82–90
- Sanchez-Inchausti G, Vaquero-Martin J, Vidal-Fernandez C. Effect of arthroscopy and continuous cryotherapy on the intra-articular temperature of the knee. Arthroscopy 2005;21:552–56
- Ito A, Aoyama T, Iijima H, Nagai M, Yamaguchi S, Tajino J, et al. Optimum temperature for extracellular matrix production by articular chondrocytes. Int J Hyperthermia 2014;30:96–101
- Serrat MA, King D, Lovejoy CO. Temperature regulates limb length in homeotherms by directly modulating cartilage growth. Proc Natl Acad Sci USA 2008;105:19348–53
- Falk MH, Issels RD. Hyperthermia in oncology. Int J Hyperthermia 2001;17:1–18
- Takahashi KA, Tonomura H, Arai Y, Terauchi R, Honjo K, Hiraoka N, et al. Hyperthermia for the treatment of articular cartilage with osteoarthritis. Int J Hyperthermia 2009;25:661–7
- McArdle A, Dillmann WH, Mestril R, Faulkner JA, Jackson MJ. Overexpression of HSP70 in mouse skeletal muscle protects against muscle damage and age-related muscle dysfunction. FASEB J 2004;18:355–7
- Terauchi R, Takahashi KA, Arai Y, Ikeda T, Ohashi S, Imanishi J, et al. Hsp70 prevents nitric oxide-induced apoptosis in articular chondrocytes. Arthritis Rheum 2003;48:1562–8
- Beckham JT, Wilmink GJ, Mackanos MA, Takahashi K, Contag CH, Takahashi T, et al. Role of HSP70 in cellular thermotolerance. Lasers Surg Med 2008;40:704–15
- Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalisation: Strategies and considerations. Genes Immun 2005;6:279–84
- Guenin S, Mauriat M, Pelloux J, Van Wuytswinkel O, Bellini C, Gutierrez L. Normalization of qRT-PCR data: The necessity of adopting a systematic, experimental conditions-specific validation of references. J Exp Bot 2009;60:487–93
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002;3:0034.1–11
- Foldager CB, Munir S, Ulrik-Vinther M, Soballe K, Bunger C, Lind M. Validation of suitable housekeeping genes for hypoxia-cultured human chondrocytes. BMC Mol Biol 2009;10:94
- Ito A, Aoyama T, Yamaguchi S, Zhang X, Akiyama H, Kuroki H. Low-intensity pulsed ultrasound inhibits messenger RNA expression of matrix metalloproteinase-13 induced by interleukin-1beta in chondrocytes in an intensity-dependent manner. Ultrasound Med Biol 2012;38:1726–33
- Dewey WC, Hopwood LE, Sapareto SA, Gerweck LE. Cellular responses to combinations of hyperthermia and radiation. Radiology 1977;123:463–74
- Wheatley DN, Kerr C, Gregory DW. Heat-induced damage to HeLa-S3 cells: Correlation of viability, permeability, osmosensitivity, phase-contrast light-, scanning electron- and transmission electron-microscopical findings. Int J Hyperthermia 1989;5:145–62
- Pelttari K, Steck E, Richter W. The use of mesenchymal stem cells for chondrogenesis. Injury 2008;39:S58–65
- Caron MM, Emans PJ, Coolsen MM, Voss L, Surtel DA, Cremers A, et al. Redifferentiation of dedifferentiated human articular chondrocytes: Comparison of 2D and 3D cultures. Osteoarthritis Cartilage 2012;20:1170–78
- Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 2004;64:5245–50
- Studer D, Lischer S, Jochum W, Ehrbar M, Zenobi-Wong M, Maniura-Weber K. Ribosomal protein l13a as a reference gene for human bone marrow-derived mesenchymal stromal cells during expansion, adipo-, chondro-, and osteogenesis. Tissue Eng Part C Methods 2012;18:761–71
- Curtis KM, Gomez LA, Rios C, Garbayo E, Raval AP, Perez-Pinzon MA, et al. EF1alpha and RPL13a represent normalization genes suitable for RT-qPCR analysis of bone marrow derived mesenchymal stem cells. BMC Mol Biol 2010;11:61
- Walker NJ. Tech.sight.featuring real-time PCR. A technique whose time has come. Science 2002;296:557–9
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol Lett 2004;26:509–15
- Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun 2004;313:856–62
- Peters IR, Peeters D, Helps CR, Day MJ. Development and application of multiple internal reference (housekeeper) gene assays for accurate normalisation of canine gene expression studies. Vet Immunol Immunopathol 2007;117:55–66
- Coyle CH, Izzo NJ, Chu CR. Sustained hypoxia enhances chondrocyte matrix synthesis. J Orthop Res 2009;27:793–9
- Domm C, Schunke M, Christesen K, Kurz B. Redifferentiation of dedifferentiated bovine articular chondrocytes in alginate culture under low oxygen tension. Osteoarthritis Cartilage 2002;10:13–22
- Brighton CT, Heppenstall RB. Oxygen tension in zones of the epiphyseal plate, the metaphysis and diaphysis. An in vitro and in vivo study in rats and rabbits. J Bone Joint Surg Am 1971;53:719–28
- Pfander D, Gelse K. Hypoxia and osteoarthritis: How chondrocytes survive hypoxic environments. Curr Opin Rheumatol 2007;19:457–62