ABSTRACT
Background. MicroRNAs (miRNAs) have been associated with prognosis in esophageal cancer, suggesting a role for miRNAs to help guide treatment decisions. Especially, miR-21 and miR-375 have been investigated as prognostic biomarkers. The aim of this study was to evaluate the prognostic potential of miR-21 and miR-375 in primary esophageal squamous cell carcinomas (ESCC) and esophagogastric adenocarcinomas (EAC).
Material and methods. Pre-therapeutic tumor specimens from 195 patients with loco-regional esophageal cancer treated with neoadjuvant or definitive chemoradiotherapy or perioperative chemotherapy were analyzed. Expression levels of miR-21 and miR-375 were quantified using Affymetrix GeneChip miRNA 1.0 Array. The Cox proportional hazards model was used to assess the correlation of miR-21 and miR-375 with disease-specific survival (DSS) and overall survival (OS). Forest plots were performed to evaluate the prognostic impact of miR-21 and miR-375 in the present study and previously published reports.
Results. In ESCC, patients with miR-21 expression levels above median showed a trend towards poorer DSS and OS. When dividing miR-21 expression by tertiles, high levels of miR-21 significantly correlated with shortened DSS [HR 1.76 (95% CI 1.05–2.97) but not OS. Similarly for EAC, a significant association between miR-21 expression above median and DSS was observed [HR 3.37 (95% CI 1.41–8.05)], in addition to a trend towards poorer OS for patients with miR-21 expression above median. Multivariate analyses identified miR-21 as an independent prognostic marker for DSS in EAC [HR 3.52 (95% CI 1.06–11.69)]. High miR-375 was not correlated with improved prognosis in either histology. However, Forest plots demonstrated that both miR-21 and miR-375 were of prognostic impact in ESCC.
Conclusion. In this study, miR-21 was identified as an independent prognostic biomarker for DSS in patients with EAC whereas miR-21 failed to show independent prognostic significance in ESCC. High miR-375 was not associated with enhanced survival in either histology.
Esophageal cancer (EC) is a group of heterogeneous malignancies with squamous cell carcinoma (ESCC) and adenocarcinoma (EAC) as the most prevalent histological subtypes. Despite significant therapeutic improvements, the five-year overall survival (OS) rate ranges from 15% to 25% [Citation1]. EC is often diagnosed at an advanced stage, requiring multi-modality t herapy including surgery, chemotherapy and irradiation. Such comprehensive treatment strategies are potentially toxic and might be associated with severe side effects. Unfortunately, a substantial amount of patients will gain no benefit from these standard treatment regimens. Hence, pre-therapeutic prognostic factors are crucial in order to improve patient selection and clinical management of the disease.
MicroRNAs (miRNAs) are well conserved endogenous, small, non-coding RNAs (18–24 nucleotides) that mainly bind imperfectly to the 3’ untranslated region of target messenger RNAs (mRNA) [Citation2]. They act as post-transcriptional regulators by targeting mRNA for degradation or translational repression, usually resulting in gene silencing [Citation2]. miRNAs are involved in regulation of many cellular processes, including proliferation, apoptosis and differentiation and are key players in cancer pathogenesis, functioning as tumor suppressors or oncogenes [Citation2,Citation3]. In addition, miRNA expression profiling has shown an association between miRNAs and clinical outcome, indicating the use of miRNAs to help identify low-risk patients from high-risk patients and thereby to help guide treatment decisions.
miR-21 stands out as the most frequently overexpressed miRNA in cancers, e.g. lung cancer, breast cancer, colorectal cancer and pancreatic cancer [Citation4], and is suggested to serve as a prognostic biomarker for human malignancies [Citation5]. miR-21 is known as an oncogenic miRNA (oncomir), targeting several tumor-suppressor genes, resulting in increased tumor growth, invasion, metastasis and reduced sensitivity to chemotherapy [Citation6–9]. Also in EC, miR-21 has been evaluated for its prognostic potential in both ESCC and EAC [Citation7,Citation10–15].
In addition, the tumor suppressor, miR-375, has been studied as a prognostic biomarker in EC [Citation12,Citation13,Citation16–18]. miR-375 is most often down-regulated in EC and is known to serve anti-oncogenic properties, attributing to suppressed tumor proliferation, inhibited tumor growth and decreased metastasis rate [Citation19]. The observed associations on both miR-21 and miR-375 have been discordant with some studies showing significant associations with patient survival and others insignificant results. However, in meta-analyses, miR-21 and miR-375 have been identified as promising prognostic biomarkers in EC [Citation20,Citation21], showing association with outcome. Hence, elevated miR-21 levels and reduced miR-375 levels have been significantly associated with poorer prognosis, primarily in ESCC. Nevertheless, these results were not conclusive as the analyses suffered from lack of eligible studies, limited sample sizes and contradicting findings.
In order to test these observations, the current study was performed based on a retrospective study population and with the aim to evaluate the prognostic impact of miR-21 and miR-375 in two cohorts of patients with ESCC and EAC, respectively.
Material and methods
Patient and tumor characteristics
The retrospective study population comprised 195 patients with loco-regional gastroesophageal cancer (GEC) with available formalin-fixed, paraffin- embedded (FFPE) tumor specimens with sufficient RNA for microarray analysis. All specimens were pre-therapeutic biopsies from previously untreated, newly diagnosed patients with GEC. Patients had undergone curatively intended therapy at three independent clinical centers in Denmark: 1) Department of Oncology, Aarhus University Hospital, 2) Department of Oncology, Odense University Hospital, and 3) Department of Oncology, Rigshospitalet, Copenhagen. Clinicopathological parameters were obtained from medical records including pathology reports. The study was approved by The Central Denmark Region Committees on Health Research Ethics and the Danish Data Protection Agency and was conducted in accordance with the Helsinki declaration.
ESCC and EAC are heterogeneous malignancies with different etiologies, locations and treatment strategies. In addition, miRNAs are known to serve tumor-type specificity. Hence, the present study population was divided and studied on the basis of histology. ESCC was diagnosed in 129 patients (66%) and adenocarcinoma of the esophagus/esophagogastric junction (N = 61) or stomach (N = 5) in 66 patients (34%). From here on patients with adenocarcinoma will be referred to as EAC and the entire study cohort as diagnosed with EC. Baseline patient and tumor characteristics are summarized in . Briefly, for ESCC patients, the median age at diagnosis was 63 years (range 36–81 years) with a male to female ratio of 85:44. The predominant cT-stage and cN-stage was cT3 and cN1, respectively. Neoadjuvant chemoradiotherapy (concurrent 5-FU with/without cisplatin and 45–50 Gy) was given to 90 patients (70%) and definitive chemoradiotherapy (concurrent cisplatin, 5-FU and 50–60 Gy) to 39 patients (30%).
Table I. Patient and tumor characteristics.
For EAC, the median age was 64 years (range 32–86 years), male to female ratio 55:11 and the predominant cT-stage and cN-stage was cTX, indicating that cT-stage was not available in a number of cases, and cN1. Forty-six patients (70%) received perioperative chemotherapy (cisplatin, epirubicin and capecitabine), 18 patients (27%) definitive chemoradiotherapy (concurrent cisplatin, 5-FU and 50–60 Gy) and two patients (3%) neoadjuvant chemoradiotherapy (concurrent cisplatin/5-FU and 45–50 Gy).
RNA isolation and quantification of miRNA
miRNA was isolated from five whole slides of FFPE tissue sections with a thickness of 12 μm using the RecoverAllTM Total Nucleic Acid Isolation Kit for FFPE (Ambion, Austin, TX, USA) according to the manufacturer protocol. Hematoxylin and eosin-stained slides were used to verify the presence of invasive carcinoma and confirmation of tumor presence was carried out by an experienced pathologist. The mean fraction of tumor area (defined as the area of invasive carcinoma) was estimated to 63% (range 10–100). Total RNA was quantified with a NanoDrop spectrophotometer (NanoDrop Technology, Wilmington, DE, USA).
Analysis of miRNAs was performed using Affymetrix GeneChip miRNA microarrays comprising 847 human miRNAs probe sets. miRNAs were labeled using FlashTagTM Biotin HSR RNA Labeling Kit (Affymetrix, Santa Clara, CA, USA) and hybridized to GeneChip miRNA version 1.0 microarrays (Affymetrix) according to manufacturer's details. Arrays were washed and stained on an Affymetrix Fluidics Station 450X and scanned on an Affymetrix G7 scanner according to manufacturer's instructions. Normalization of miRNA microarray expression values were performed in R using Robust Multi-array Average (RMA). miR-21 and miR-375 expression levels were measurable in all 195 tumor samples.
Treatment outcomes
Patients were followed from the date of histological diagnosis until death or last day of follow-up (11 August 2014). All patients were followed for at least 20 months or until death.
The primary outcome was disease-specific survival (DSS), defined as time from diagnosis to death from or with EC or last follow-up. Secondary outcomes were OS, defined as the time interval from diagnosis of EC to death from any cause or last follow-up; pathological complete response (ypCR), defined as patients with no evidence of vital residual tumor cells remaining in the esophagectomy specimen after neoadjuvant treatment and radiographic response, defined as patients with complete response or regression of disease.
Pathological response evaluation was performed by experienced pathologists subspecialized in gastrointestinal pathology. Radiographic response was assessed in patients with treatment evaluation CT scans after completion of induction therapy prior to surgery.
Statistics
For each cohort (ESCC and EAC), survival analysis was carried out by dichotomizing the miRNA expression values using the median expression as a cut-off or by division of miRNA expression into tertiles. Survival estimates were calculated according to the Kaplan-Meier method and compared using the univariate Cox proportional hazards model to assess associations between miRNA expression levels and DSS and OS. The assumption of proportional hazards was verified with Schoenfeld's residuals. In addition, multivariate analysis was performed to adjust for the clinicopathological parameters age, sex, cT- and cN-stage. Hazard ratios (HRs) for both uni- and multivariate analyses are presented at 3 years with 95% confidence interval (CI). Association of miRNA expression and treatment response was carried out using the χ2 statistical test or Fisher's exact test. Forest plots were performed to evaluate the association of the prognostic role of miR-21 and miR-375 in the present study and previously published reports. If only results from log-rank tests were provided in published studies, HRs and the corresponding 95% CIs were estimated using the p-value and number of events (extracted from risk-tables or Kaplan-Meier curves) as previously described by Tierney et al. [Citation22]. Statistical significance was defined as two-sided p < 0.05. All statistical analyses were performed using STATA, Version 12 (StataCorp, College Station, TX, USA).
Results
Outcome analysis
ESCC
The median follow-up time was 24 months (3–192 months), with 67% (59–75) of patients surviving at one year and 40% (31–48) at three years. Pathological complete response was observed in 28 patients (41%) of 68 undergoing esophagectomy and radiographic response in 77 patients (63%) of 122 patients with available treatment evaluation CT scans.
Univariate Cox proportional hazards model indicated that cT-stage was significantly correlated with both DSS [1.99 (95% CI 1.03–3.86)] and OS [1.78 (95% CI 1.01–3.16)], and cN-stage to be significantly associated with DSS [2.16 (95% CI 1.09–4.28)] (). However, neither cT- nor cN-stage were independent prognostic markers in a multivariate analysis ().
Table II. Uni- and multivariate analyses.
EAC
Median follow-up time was 23 months (4–99 months). The one-year survival rate was 77% (65–86) and the three-year survival rate 46% (33–58). No patients obtained pathological complete response. Radiographic response was observed in 37 patients (61%) of 61 patients with evaluation CT scans.
For patients with EAC, cT-stage was observed to be significantly associated with OS [2.50 (95% CI 1.01–6.22)], but not DSS (). In a multivariate model, however, cT-stage was not identified to be an independent prognostic factor for OS (). Due to limited number of events and insignificant estimates of age and sex in the univariate analysis, only cT- and cN-stage was included in the multivariate analysis of EAC.
Prognostic value of miR-21 and miR-375 in ESCC
In order to assess the potential prognostic impact of miR-21 and miR-375, miRNA expression values derived from microarray analysis were dichotomized into groups of high and low expression based on a median cut-off for each miRNA. Univariate analysis showed a trend towards a poorer DSS for ESCC patients with miR-21 expression above the median cut-off [HR 1.28 (95% CI 0.77–2.15)] (). Similarly, when evaluating OS a worse outcome was observed in the group of patients with miR-21 expression above the median [HR 1.26 (95% CI 0.80–1.98)] (). To further elucidate the correlation of miR-21 and prognosis, expression levels were divided into tertiles. The tertile including patients with highest miR-21 expression values (upper tertile) was considered against the other two tertiles as these two tertiles were overlapping, suggesting no difference in survival between patients with intermediate and lower miR-21 expression. As shown in and D, high miR-21 expression was significantly associated with shortened DSS OS [HR 1.76 (95% CI 1.05–2.97) whereas only a trend towards a poorer OS was observed for patients with high miR-21 expression [HR: 1.50 (95% CI 0.95–2.39)]. A multivariate analysis including age, sex, T- and N-stage did not identify miR-21 as an independent prognostic marker for neither DSS nor OS (divided by tertiles) ().
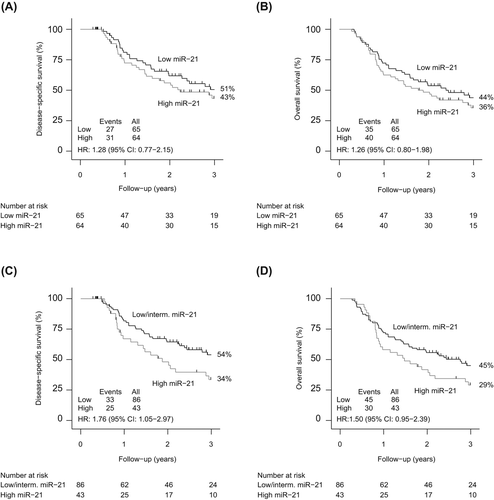
No associations between high expression levels of miR-375 and DSS or OS were identified in patients with ESCC when using the median expression level as cut-off [HR 0.82 (95% CI 0.49–1.38) and HR 0.88 (95% CI 0.56–1.39)]. Similarly, when dividing expression levels by tertiles, no prognostic value of miR-375 was observed.
Prognostic value of miR-21 and miR-375 in EAC
Similar to the analysis of ESCC, median expression was used as cut-off in the group of EAC patients. A significant correlation between high miR-21 expression and DSS was observed for patients with EAC [HR 3.37 (95% CI 1.41–8.05)] (). In addition, a trend towards a poorer OS was shown for patients with induced miR-21 expression [HR 1.96 (95% CI 0.98–3.94)] (). Next, expression levels were divided into tertiles. No significant association between outcome and miR-21 expression levels were observed [DSS: Intermediate vs. low expression: HR 2.08 (95% CI: 0.71–6.08) and high vs. low expression: HR 2.28 (95% CI: 0.79–6.59) and OS: Intermediate vs. low expression: HR 1.04 (95% CI: 0.45–2.39) and high vs. low expression: HR 1.13 (95% CI: 0.50–2.57)] ( and D). In a multivariate analysis, miR-21 was identified to an independent prognostic marker for DSS [HR 3.52 (95% CI: 1.06-11.69)] but not for OS (divided by median) ().
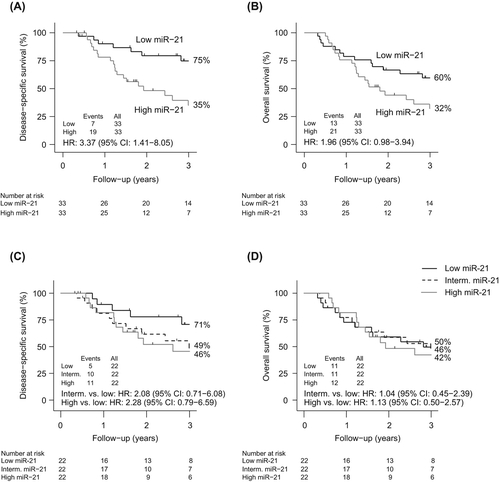
Evaluation of miR-375 in EAC patients showed no correlations with DSS or OS when using the median expression level as cut-off [HR 1.23 (95% CI 0.57–2.66) and HR 1.53 (95% CI 0.78–3.02)]. When dividing expression levels by tertiles, no correlation between high miR-375 expression and improved survival was observed.
Correlation of miR-21, miR-375 and treatment response
Using the χ2 statistical test or Fisher's exact test, miR-21 expression was not associated with pathological response in patients with ESCC (P=0.154). Radiographic response, however, was significantly correlated with miR-21 expression (P=0.049) in that low miR-21 expression was correlated to non-radiographic response. miR-375 expression was not associated with neither pathological or radiographic response (P=0.931 and P=0.573, respectively) in patients with ESCC. Similarly, for EAC, no association between miR-21 expression and radiographic response was observed (P=0.404) or miR-375 and radiographic response (P=0.192).
miR-21 and miR-375 as prognostic markers in ESCC and EAC (comparing present and previous publications by Forest plot analyses)
Forest plots were performed in order to study the association between the present results in context with previously published reports on the prognostic role of miR-21 and miR-375 in ESCC. For studies evaluating miR-21, four studies were included in the analysis [Citation12–15]. These studies either provided HRs and 95% CIs directly [Citation13] or described log-rank tests including number of events in which it was possible to calculate the corresponding HRs and their 95% CIs [Citation12,Citation14,Citation15]. Three studies reported no statistical significant association between miR-21 expression levels in cancerous tissue and survival [Citation12–14] whereas only one study showed a significant and inverse correlation between miR-21 and OS [Citation15]. However, despite these discordant results, a Forest plot identified a pooled HR of 1.56 (95% CI 1.18–2.07), indicating that high miR-21 expression level significantly predicted a poorer survival (). For miR-375, the HRs and their 95% CIs were assessed in five studies [Citation12,Citation13,Citation16–18]. Of these, two studies described no significant association between miR-375 expression and prognosis [Citation12,Citation13] and three studies a significant association with survival [Citation16–18]. A Forest plot showed a statistical significant correlation between up-regulated miR-375 expression and increased survival in ESCC, with a pooled HR of 0.62 (95% CI 0.49–0.79) ().
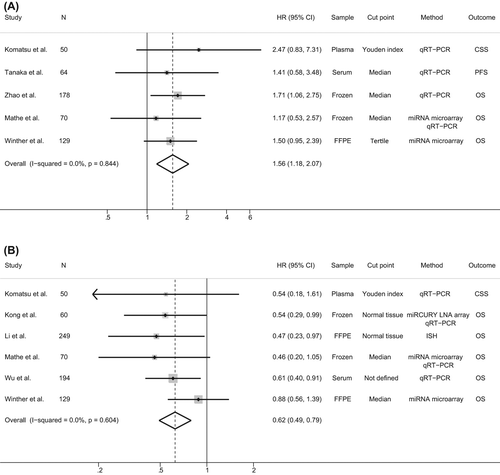
As HRs and 95% CIs were only assessed in two studies evaluating miR21 in EAC and one study evaluating miR-375 in EAC, pooled analysis was not performed.
Discussion
To the best of our knowledge, this is the largest study to date to assess the potential prognostic value of miR-21 and miR-375 in EC in a Western population. These two candidate miRNAs were evaluated in retrospective cohorts of 129 patients with ESCC and 66 patients with EAC. In the present study, miR-21 expression was significantly correlated with DSS in patients with ESCC and EAC. In addition, miR-21 was identified as an independent prognostic biomarker in EAC but not ESCC. However, a Forest plot including the present result showed that miR-21 was indeed a poor prognostic marker in ESCC. miR-21 is a known oncomir, implicated in tumor pathogenesis and carcinogenesis. Several studies have shown that miR-21 targets and down-regulates tumor suppressor genes, such as phosphatase and tensin homolog (PTEN) [Citation7,Citation23], programmed cell death 4 (PDCD4) [Citation8], FASL, TIMP3 and RECK [Citation9], which are involved in tumorigenesis, apoptosis, invasion and metastasis. In addition, miR-21 has been subjected to substantial investigation of its association with clinical outcome and emerging evidence indicates that miR-21 might have prognostic potential.
When evaluating miR-375 in this study population, no significant correlations between high miR-375 expression and increased DSS or OS were observed for neither ESCC nor EAC. However, comparison with previous published studies identified a significant association between high miR-375 expression and prolonged survival in ESCC. miR-375 is an acknowledged tumor suppressor, frequently down-regulated in EC due to hypermethylation of the promotor. It has been demonstrated that miR-375 negatively regulates 3-phosphoinositide-dependent protein kinase-1 (PDK1) [Citation24] and insulin-like growth factor 1 receptor (IGF1R) [Citation16], attributing to suppressed tumor proliferation and metastasis. This is consistent with previous reports on associations between repressed miR-375 expression levels and poor prognosis in ESCC [Citation16–18]. Similarly, for EAC, Mathe et al. found that low expression of miR-375 was significantly correlated with worse prognosis, but only in EAC patients with Barrett's esophagus [Citation13].
The clinical rational for using prognostic biomarkers, such as miRNAs, is the potential to predict clinical outcome, enabling clinicians to identify high-risk patients who might benefit from more aggressive treatment strategies and to identify low-risk patients who might be safely treated with less intensive treatment regiments and, thereby, reduce toxic side effects in this group of patients. Several studies have investigated the prognostic value of miR-21 and miR-375 expression levels in EC. However, results have been contradicting. In this study, comparison of previously published studies using Forest plots suggested that miR-21 was significantly and inversely correlated with survival and that high miR-375 expression level was significantly associated with enhanced survival of patients with ESCC. Despite these intriguing results, the conclusions might be biased by several factors. First, the number of studies included in the analyses was limited to eight including the present study and sample sizes were small. For ESCC the median sample size was 70 (range 50–178) and for miR-375 the median sample size was 68 (range 50–249). Second, miRNA expression levels were obtained from different origins including FFPE tumor specimens, frozen tumor tissues, serum and plasma. In addition, miRNA expression levels were obtained using different methodologies, such as miRNA microarray, in situ hybridization (ISH) and qRT-PCR. Third, endpoints differed between studies [OS, progression-free survival (PFS) and cause-specific survival (CSS)] with OS being the most commonly used parameter. Also, it should be mentioned that most of the HRs and CIs in the present analyses were generated from log-rank test and calculation of events, indicating that the estimates might be subjected to some uncertainty.
Furthermore, and of potentially crucial impact for interpretation of results, cut-off definitions of miRNA expression levels varied between studies. Splitting miRNA expression levels by the sample median was the choice of preference in three studies evaluated in the present analyses with Forest plots [Citation13–15]. Other studies defined the cut-offs using Youden index (N = 1) [Citation12] or normal tissue (N = 2) [Citation16,Citation17]. One study did not define the cut-off [Citation18]. In our study, a cut-off using the median miRNA expression value was chosen, in addition to, a cut-off using tertiles in order to explore the correlation of miRNA expression and prognosis further.
Besides evaluation of the prognostic impact of miR-21 and miR-375, the present study also investigated the association between the two miRNAs and treatment response in terms of pathological or radiographic response. However, neither miR-21 nor miR-375 expression was associated with pathological and/or radiographic response in patients with ESCC or EAC. In previous reports, serum miR-21 expression levels have been shown to be significantly decreased in ESCC patients achieving complete or partial response after chemotherapy compared to patients with stable disease or progressive disease [Citation25]. In addition, in vitro studies have demonstrated that overexpression of miR-21 in TE-1 cells correlated with radioresistance and that inhibition of miR-21 increased radiosensitivity, possible through activation of PTEN [Citation23]. Studies on miR-375 and treatment response in EC are yet to be performed. Despite, the preliminary nature of these results, the findings demonstrate the potential role of miRNAs as biomarkers of treatment response and the importance of identifying miRNAs that would help predict treatment outcome.
In conclusion, esophageal cancers are heterogeneous malignancies and the prognostic impact of miR-21 and miR-375 was evaluated in cohorts of ESCC and EAC. miR-21 was identified as an independent prognostic biomarker for DSS in EAC, but not in ESCC. However, no significant correlations between high miR-375 expression and improved prognosis were observed in neither ESCC nor EAC.
Acknowledgments
The authors thank Mogens Jøns Johannsen for excellent technical assistance and Medical Prognosis Institute for performing microarray analysis. The study was financially supported by The Danish Cancer Society, CIRRO – The Lundbeck Foundation Center for Interventional Research in Radiation Oncology and The Danish Council for Strategic Research, and Karen A. Tolstrups Fund.
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet 2013;381:400–12.
- Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer 2006;6:259–69.
- Kent OA, Mendell JT. A small piece in the cancer puzzle: MicroRNAs as tumor suppressors and oncogenes. Oncogene 2006;25:6188–96.
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006;103:2257–61.
- Zhou X, Wang X, Huang Z, Wang J, Zhu W, Shu Y, et al. Prognostic value of miR-21 in various cancers: An updating meta-analysis. PLoS One 2014;9:e102413.
- Seca H, Lima RT, Lopes-Rodrigues V, Guimaraes JE, Almeida GM, Vasconcelos MH. Targeting miR-21 induces autophagy and chemosensitivity of leukemia cells. Curr Drug Targets 2013;14:1135–43.
- Li P, Mao WM, Zheng ZG, Dong ZM, Ling ZQ. Down-regulation of PTEN expression modulated by dysregulated miR-21 contributes to the progression of esophageal cancer. Dig Dis Sci 2013;58:3483–93.
- Hiyoshi Y, Kamohara H, Karashima R, Sato N, Imamura Y, Nagai Y, et al. MicroRNA-21 regulates the proliferation and invasion in esophageal squamous cell carcinoma. Clin Cancer Res 2009;15:1915–22.
- Wang N, Zhang CQ, He JH, Duan XF, Wang YY, Ji X, et al. MiR-21 down-regulation suppresses cell growth, invasion and induces cell apoptosis by targeting FASL, TIMP3, and RECK genes in esophageal carcinoma. Dig Dis Sci 2013;58:1863–70.
- Hu Y, Correa AM, Hoque A, Guan B, Ye F, Huang J, et al. Prognostic significance of differentially expressed miRNAs in esophageal cancer. Int J Cancer 2011;128:132–43.
- Hummel R, Hussey DJ, Michael MZ, Haier J, Bruewer M, Senninger N, et al. MiRNAs and their association with locoregional staging and survival following surgery for esophageal carcinoma. Ann Surg Oncol 2011;18:253–60.
- Komatsu S, Ichikawa D, Takeshita H, Konishi H, Nagata H, Hirajima S, et al. Prognostic impact of circulating miR-21 and miR-375 in plasma of patients with esophageal squamous cell carcinoma. Expert Opin Biol Ther 2012; 12(Suppl 1):S53–9.
- Mathe EA, Nguyen GH, Bowman ED, Zhao Y, Budhu A, Schetter AJ, et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: Associations with survival. Clin Cancer Res 2009; 15:6192–200.
- Tanaka K, Miyata H, Yamasaki M, Sugimura K, Takahashi T, Kurokawa Y, et al. Circulating miR-200c levels significantly predict response to chemotherapy and prognosis of patients undergoing neoadjuvant chemotherapy for esophageal cancer. Ann Surg Oncol 2013;20(Suppl 3):S607–15.
- Zhao Y, Schetter AJ, Yang GB, Nguyen G, Mathe EA, Li P, et al. microRNA and inflammatory gene expression as prognostic marker for overall survival in esophageal squamous cell carcinoma. Int J Cancer 2013;132:2901–9.
- Kong KL, Kwong DL, Chan TH, Law SY, Chen L, Li Y, et al. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut 2012; 61:33–42.
- Li J, Li X, Li Y, Yang H, Wang L, Qin Y, et al. Cell-specific detection of miR-375 downregulation for predicting the prognosis of esophageal squamous cell carcinoma by miRNA in situ hybridization. PLoS One 2013;8:e53582.
- Wu C, Li M, Hu C, Duan H. Clinical significance of serum miR-223, miR-25 and miR-375 in patients with esophageal squamous cell carcinoma. Mol Biol Rep 2014; 41:1257–66.
- Yan JW, Lin JS, He XX. The emerging role of miR-375 in cancer. Int J Cancer 2014;135:1011–8.
- Fu C, Dong W, Wang Z, Li H, Qin Q, Li B. The expression of miR-21 and miR-375 predict prognosis of esophageal cancer. Biochem Biophys Res Commun 2014;446: 1197–203.
- Fu W, Pang L, Chen Y, Yang L, Zhu J, Wei Y. The microRNAs as prognostic biomarkers for survival in esophageal cancer: A meta-analysis. Sci World J 2014;2014:523979.
- Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 2007;8:16.
- Huang S, Li XQ, Chen X, Che SM, Chen W, Zhang XZ. Inhibition of microRNA-21 increases radiosensitivity of esophageal cancer cells through phosphatase and tensin homolog deleted on chromosome 10 activation. Dis Esophagus 2013;26:823–31.
- Li X, Lin R, Li J. Epigenetic silencing of microRNA-375 regulates PDK1 expression in esophageal cancer. Dig Dis Sci 2011;56:2849–56.
- Kurashige J, Kamohara H, Watanabe M, Tanaka Y, Kinoshita K, Saito S, et al. Serum microRNA-21 is a novel biomarker in patients with esophageal squamous cell carcinoma. J Surg Oncol 2012;106:188–92.
