Abstract
Background: Phospholipase A2 Group IV C (PLA2G4C) catalyzes the release of certain fatty acids from phospholipids and plays a role in a range of physiological functions, such as remodeling of cell membranes and the production of prostaglandins. Furthermore, it has been proposed that PLA2G4C plays an important role in breast cancer cell chemotaxis. This study aimed to investigate the effect of a single nucleotide polymorphism (SNP) rs1549637 (T>A) of the PLA2G4C gene on the prognosis of colorectal cancer (CRC).
Material and methods: Whole blood DNA was extracted from 381 patients with CRC and 618 controls, and a TaqMan SNP genotyping assay was used to determine the distribution of the genotypes. Cancer-specific and disease-free survival was analyzed by Kaplan-Meier graphs and by uni- and multivariable Cox regression.
Results: The cancer-specific survival differed between the genotypes (p = 0.019) and the carriers of the A allele were associated with the highest risk of CRC death, with a hazard ratio (HR) of 1.72 [95% confidence interval (CI) 1.17–2.53, p = 0.006] compared with homozygous carriers of the T allele. This increased mortality in the carriers with the allele A was especially marked in stage II with an HR of 3.84 (95% CI 1.51–9.78, p = 0.005).
Conclusion: The A allele in PLA2G4C SNP (rs1549637) is associated with a worse prognosis in patients with CRC, especially in stage II disease, and it could be a potential prognostic biomarker in the planning of individual adjuvant therapy in stage II patients.
Colorectal carcinogenesis is a multistep process maintained by accumulation of genetic and epigenetic aberrations in several pathways [Citation1–3]. These include the chromosomal instability pathways, which are associated with accumulation of activating mutations in oncogenes, such as KRAS, BRAF and PI3KCA, or inactivation of tumor suppressor genes, such as APC and P53 [Citation1,Citation2]. Moreover, epigenetic events, such as aberrant methylation of gene promoter regions, affect pathways in colorectal cancer (CRC) initiation and progression [Citation3,Citation4].
The expression of the enzyme cyclooxygenase 2 (COX-2) is elevated in CRC [Citation5–7] and its proinflammatory and protumorigenic roles are mediated by prostaglandins, such as mitogenic prostaglandin E2 (PGE2), which has an anti-apoptotic effect [Citation8,Citation9]. In brief, lipid metabolism, especially the arachidonic acid (AA) metabolizing pathway, plays a role in chronic inflammation and colorectal carcinogenesis [Citation10]. The phospholipase A2 (PLA2) family consists of enzymes defined by their ability to catalyze fatty acid from the sn-2 position of glycerophospholipids [Citation11,Citation12]. The hydrolysis product of the PLA2 reaction is a free fatty acid, such as AA. Subsequently, AA will serve as a substrate for COX-2 which generates eicosanoids, such as PGE2 [Citation12,Citation13]. Among the forms of PLA2s expressed in human tissue, phospholipase A2 Group IV A (PLA2G4A), so-called, 85 kDa cytosolic PLA2, has been shown to be overexpressed in CRC [Citation7,Citation14–16]. Interestingly, another form of PLA2, PLA2G2A seems to be a prognostic determinant in selecting patients at high risk for recurrent disease in stage II who might benefit from adjuvant therapy [Citation17].
Cytosolic PLA2G4C contributes to cellular AA metabolism and phospholipid remodeling [Citation12,Citation18]. AA released by PLA2G4C can be linked to the COX-2 pathway to produce prostaglandins [Citation18]. PLA2G4C is predominantly expressed in the brain, heart and skeletal muscle in humans [Citation19]. Moreover, PLA2G4C has been reported to be located in endoplasmatic reticulum and mitochondria and to have lysophospholipase and transacylation activity beside phospholipase A2 activity [Citation19,Citation20]. The exact role of PLA2G4C expression in tumorigenesis is still unknown but it seems to play a role in breast cancer cell chemotaxis and invasion through regulating the Akt pathway [Citation21].
Genetic variations, such as single nucleotide polymorphisms (SNPs), are thought to play a role in individual variation in CRC susceptibility [Citation22,Citation23]. Polymorphic variants of genes play a role as factors that mediate inflammatory response and favor CRC progression [Citation24].
A SNP, rs1549637 (T>A) of the PLA2G4C gene has been suggested to be involved in the susceptibility to schizophrenia [Citation25,Citation26]. This SNP may exert an impact on the metabolism of membrane phospholipids. To the best of our knowledge, available data about this SNP and its importance in colorectal carcinogenesis is limited.
In the present study, the SNP rs1549637 was analyzed to assess its value as a potential risk factor and as a predictor of disease outcome in patients with CRC. We also performed immunohistochemistry to detect PLA2G4C protein expression in CRC tissue.
Material and methods
Patients and controls
This prospective study included 381 patients (213 males and 168 females) with a mean age of 70 years (range 25–94 years), from southeastern Sweden who underwent surgical resection for primary colorectal adenocarcinomas at the Department of Surgery, Ryhov County Hospital, Jönköping, Sweden between 1996 and 2013.
Clinical information about preoperative and postoperative staging, localization of tumor, histopathological characteristics, any adjuvant chemotherapy and follow-up information about date of recurrence or death related to CRC, was obtained from the computerized patients’ files which cover all the healthcare providers in the region. Follow-up ended on the date of death or 23 February 2015. The key clinicopathological data of the patients are summarized in .
Table I. Patients’ characteristics and association with the genotypic distribution of PLA2G4C gene polymorphism (rs 1549637).
Blood donors (n = 618) with no known CRC history, and who were from the same geographical region as the patients with CRC were selected as control subjects. The control group consisted of 334 males and 284 females, with a mean age of 60 years (range 33–93 years). The investigation was approved by the local Ethical Committee (Dnr. 2013/271-31; Linköping, Sweden) and an informed consent was obtained from the participants.
DNA extraction and genotype determination
Blood samples for the patients were drawn at the start of the operation and for the controls at the time of the blood donation. The samples were centrifuged to separate plasma and blood cells and then stored at −78 °C. Genomic DNA was isolated from the lymphocytes using the QiaAmp DNA Blood Kit (Qiagen, Hilden, Germany). The TaqMan SNP genotyping assay (Applied Biosystems, Foster City, CA, USA) was used for the analysis of the rs1549637 PLA2G4C genotype. DNA (10 ng) was amplified using the 7500 Fast Real-Time PCR system (Applied Biosystems) in a total volume of 12 μl containing TaqMan Universal PCR Master mix (Applied Biosystems), included in the SNP genotyping assay. Amplification was performed using an initial cycle at 50 °C for 2 min, followed by one cycle at 95 °C for 10 min and finally 40 cycles at 95 °C for 15 s and at 60 °C for 1 min. A post-PCR endpoint reading was performed on each plate. The manual calling option in the allelic discrimination application ABI PRISM 7500 SDS software, version 1.3.1 (Applied Biosystems), was then used to assign genotypes.
Immunohistochemistry
The paraffin blocks of the tumor and normal tissue samples were used from the archives at the Department of Pathology, Ryhov County Hospital, Jönköping, Sweden. PLA2G4C was stained using a standard protocol on 4 μm sections from formalin-fixed paraffin-embedded tissue blocks. In brief, sections from tumor and paired normal tissue were incubated with a primary rabbit anti-human polyclonal PLA2G4C antibody (R&D Systems Europe, Abingdon, UK) in appropriate dilution overnight at 4 °C, and then with a secondary biotinylated goat anti-rabbit IgG antibody (Vector Laboratories, Ltd., Burlingame, CA, USA). Avidin-biotin peroxidase complexes (Vector Laboratories) were added, followed by visualization with 3,3’-diaminobenzidine (Vector Laboratories). Sections were counterstained with hematoxylin (Vector Laboratories) and rehydrated before coverslips were added.
Statistical analysis
As there is no previous knowledge on the association of PLA2G4C SNP and CRC the aim is hypothesis generating. We have therefore not done any sample size calculations but used all the available and eligible patients. Differences in the frequencies of the PLA2G4C gene polymorphism between patients and the control group, as well as between clinical characteristics within the CRC subgroup were analyzed using the χ2-test or Wilcoxon rank-sum test, where appropriate. The Hardy-Weinberg equilibrium was assessed with regard to the genotypes. These analyses were performed using SPSS Statistics Software (version 19, Chicago, IL, USA). Uni- and multivariable survival analysis was performed by Cox’s regression, Kaplan-Meier analysis and the log-rank test, using STATA 13 software (StataCorp, 2013, Stata Statistical software: Release 13, College Station, TX, USA). The goodness of fit of the multivariable models was tested by Harrell’s C index. The results were considered to indicate statistical significance at p < 0.05.
Results
The tumors were located in the colon in 205 cases and in the rectum in 176 cases, and were classified according to The American Joint Committee on Cancer (AJCC) (TNM) classification system: stage I, n = 67; stage II, n = 138; stage III, n = 118; and stage IV, n = 58 (). The frequencies of the PLA2G4C gene polymorphism among patients and controls indicated no difference in the genotype distribution or allelic frequencies (). There was no evidence of a deviation from Hardy-Weinberg equilibrium among the CRC patients or control subjects (data not shown).
The genotypes were not associated with demographic characteristics, such as age and gender in the CRC patients and the controls (data not shown). Moreover, the genotypes were not associated with clinical characteristics of the patients.
Table II. Genotypic and allelic distributions in % (n) of PLA2G4C gene polymorphism (rs 1549637) in patients with CRC and in healthy control subjects.
PLA2G4C polymorphism and survival analysis
Median duration of follow-up was 4.7 (range 0.04–18.3) years, with a total of 2285 person years. In Kaplan-Meier survival analysis we found the cancer-specific survival differed between the genotypes (log rank test p = 0.019, ). As a result of the low prevalence of the A/A genotype we combined this genotype with the heterozygous A/T genotype to form one group. The carriers of the A allele were associated with the highest risk of CRC death (log rank test p = 0.006) with a hazard ratio (HR) of 1.72 [95% confidence interval (CI) 1.17–2.53, p = 0.006] compared with homozygous carriers of the T allele (). After adjustment for sex, age, stage, tumor localization, adjuvant chemotherapy, T4 tumor invasion, histopathological type and differentiation the HR was 2.31 (95% CI 1.48–3.60, p < 0.001). Moreover, compared with the T/T carriers, the carriers with the A allele had a poor prognosis in stage II disease (log rank test p = 0.002, ), with an increased risk of CRC death and an HR of 3.84 (95% CI 1.51–9.78, p = 0.005) and an HR 2.55 (95% CI 0.89–7.29) after adjustment for sex, age, stage, tumor localization, adjuvant chemotherapy, T4 tumor invasion, histopathological type and differentiation. Adjusting only for the significant covariates (e.g. age) gave an HR of 3.07, 95% CI 1.16–8.13, p = 0.024 for allele A carriers compared with the homozygote T carriers. Similar, but not statistically significant, associations were seen for the other stages. A similar, but statistically non-significant pattern of associations were also found for disease-free survival (), with HR 1.39 (95% CI 0.81–2.37, p = 0.230) in all stages after adjustment for stage, sex, age, tumor localization, adjuvant chemotherapy, T4 tumor invasion, histopathological type and degree of differentiation, and HR 1.72 (95% CI 0.69–4.24, p = 0.242) in stage II after adjustment for all the covariates.
Figure 1. Kaplan–Meier curve describing cancer-specific survival estimates among CRC patients of all TNM stages according to genotypes of the PLA2G4C gene polymorphism.
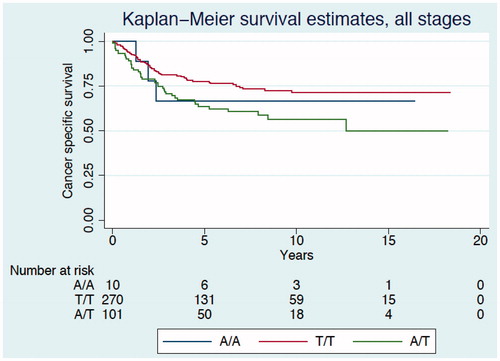
Figure 2. Kaplan–Meier curve describing cancer-specific survival estimates among CRC patients TNM stage II according to genotypes of the PLA2G4C gene polymorphism.
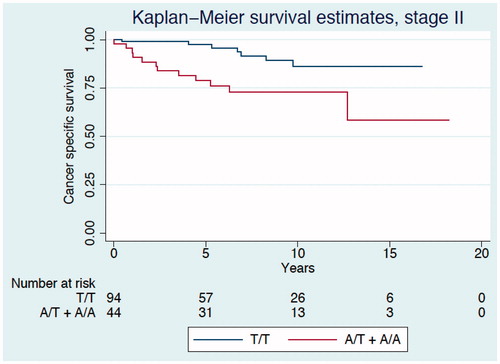
Figure 3. Kaplan–Meier curve describing disease-free survival estimates among CRC patients of all TNM stages according to genotypes of the PLA2G4C gene polymorphism.
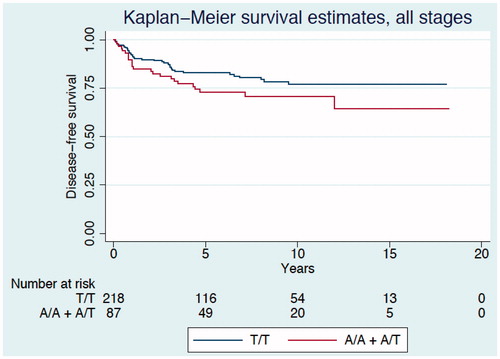
Table III. Uni-and multivariable analysis of the cancer-specific mortality.
Localization of the PLA2G4C protein in tumor and normal tissue
Expression of PLA2G4C protein was mainly localized to epithelial cells of the tumor and normal tissue. Control staining using only the secondary antibody showed no reactivity ().
Figure 4. Representative immunohistochemical staining of PLA2G4C in cancer and normal tissue from patients with CRC. The PLA2G4C protein expression is present in cancer (A) and in normal (B) epithelial cells. The arrows indicate positive brown staining. Control staining of normal tissue (C) with only the secondary antibody. Magnification, ×20.
Discussion
We found that allele A of the PLA2G4C gene is associated with a worse outcome in CRC. It remained an independent predictor of increased cancer-specific mortality also after adjustment for covariates. In stage II patients allele A was a stronger predictor for cancer-specific mortality than the conventional risk factors used for selecting stage II patients for adjuvant chemotherapy, like fewer than 12 lymph nodes removed in the specimen, T4 tumor invasion, mucinous type or histopathologic low differentiation grade, in univariable analysis. In multivariable analysis it remained among the strongest predictors, but none of the predictors reached statistical significance.
Cytosolic PLA2G4C catalyzes AA release from phospholipids and thereby contributes to cellular AA metabolism and phospholipid remodeling [Citation12,Citation18]. The AA is linked to the COX-2 pathway in the production of prostaglandins [Citation18], which exert proinflammatory, mitogenic and anti-apoptotic effects [Citation8,Citation9]. It has been suggested that an SNP, rs1549637 (T>A) of the PLA2G4C gene seems to be involved in the susceptibility to schizophrenia [Citation25,Citation26].
The present study has been done to evaluate a possible association between PLA2G4C (T>A) gene polymorphism and CRC. We observed no differences between CRC patients and controls in the genotypic and allelic frequencies and therefore the SNP seems not to be a risk factor for contracting CRC. To identify the cellular source and localization of PLA2G4C protein we used immunohistochemistry, and found expression mainly in cancer and normal epithelial cells.
Most studies of PLA2 enzymes related to CRC are concerned with expression patterns in cancer and paired normal tissue [Citation14–17]. The PLA2G2A protein expression in CRC tissue seems to be a prognostic determinant in selecting patients at high risk for recurrent disease [Citation17]. Moreover, PLA2G4C is expressed in a number of breast cancer cell lines and is suggested to play a role in cancer cell chemotaxis [Citation21].
Approximately 30% of all CRC patients are diagnosed with stage II disease. Although the prognosis is generally good for stage II disease a subgroup of patients (20–25%) will experience recurrence [Citation27–30]. Even though there is clear evidence for a benefit of adjuvant chemotherapy for stage III disease [Citation31], the potential value of such treatment for patients with stage II CRC remains controversial [Citation27–30,Citation32]. The currently used markers to select subgroups of patients with high risk for adjuvant chemotherapy (poorly differentiated tumor, lymphovascular or perineural invasion, perforation, T4 growth, elevated CEA levels and if fewer than 12 lymph nodes are removed) have all weak prognostic characteristics [Citation31]. Efforts are therefore made to find risk factors with better prognostic characteristics.
In the present study, we noted that, compared with homozygous individuals for the T allele, the carriers of the A allele of the PLA2G4C (T>A) SNP were significantly more associated with a high risk of CRC death in stage II. The genotypes A/T and A/A as markers could potentially be helpful in identifying an aggressive phenotype in stage II CRC and facilitate selection of high risk patients.
Further studies evaluating the functional properties of the investigated polymorphism rs1549637 are needed to explain whether the gene variants of this SNP differ phenotypically. Both heterozygous and homozygous gene variants could modify the properties which characterize PLA2G4C, such as PLA2, lysophospholipse and transacylation activity. In this study another point that cannot be excluded is that the investigated polymorphism may be in linkage disequilibrium with another polymorphism which is associated with clinical outcome in patients with CRC.
The strength of the study is the long follow-up period with a median follow-up time of almost five years. This may also seem like a weakness, as the management of CRC patients has gone through dramatic changes during this time. However, there is no reason that the biology of CRC have changed. Adjuvant chemotherapy may also have influenced the result, but this is less likely as the results remained almost unchanged after adjustment for adjuvant chemotherapy.
The weakness of the study is the small numbers of patients. This may explain why we failed to reach statistical significance both in the multivariable analysis of cancer-specific survival for stage II and in the analysis of the disease-free survival. However, the strength of the increased risk associated with the A allele was at the same magnitude as the other conventional risk-factors. Finally, some other risk factors, like CEA and vascular, lymphatic and neural tumor invasion was missing for the oldest part of the study period and therefore not included in the analyses.
To the best of our knowledge this is the first report on the influence of PLA2G4C gene polymorphism in patients with CRC. Our results suggest that the genetic variation of PLA2G4C may be a marker that reflects clinical outcome in patients with CRC. A possible role of the PLA2G4C gene polymorphism as a prognostic biomarker in the planning of individual therapy in stage II CRC requires further investigation.
Acknowledgements
This work was supported by grants from Futurum, the Academy of Healthcare, the Region Jönköping County, Sweden, the Foundation of Clinical Cancer Research, Jönköping Sweden and the University College of Health Sciences, Jönköping, Sweden. The authors thank Marita Skarstedt and Karin Ström for their excellent technical support.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Al-Sohaily S, Biankin A, Leong R, Kohonen-Corish M, Warusavitarne J. Molecular pathways in colorectal cancer. J Gastroenterol Hepatol 2012;27:1423–31.
- Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med 2009;361:2449–60.
- Kondo Y, Issa JP. Epigenetic changes in colorectal cancer. Cancer Metastasis Rev 2004;23:29–39.
- Naghibalhossaini F, Zamani M, Mokarram P, Khalili I, Rasi M, Mostafavi-Pour Z. Epigenetic and genetic analysis of WNT signaling pathway in sporadic colorectal cancer patients from Iran. Mol Biol Rep 2012;39:6171–8.
- Eberhart CE, Coffey RJ, Radhika A, Giardiello FM, Ferrenbach S, DuBois RN. Up-regulation of cyclooxygenase 2 gene expression in human colorectal adenomas and adenocarcinomas. Gastroenterology 1994;107:1183–8.
- Koehne CH, Dubois RN. COX-2 inhibition and colorectal cancer. Semin Oncol 2004;31:12–21.
- Dimberg J, Samuelsson A, Hugander A, Söderkvist P. Gene expression of cyclooxygenase-2, group II and cytosolic phospholipase A2 in human colorectal cancer. Anticancer Res 1998;18:3283–7.
- Sheng H, Shao J, Morrow RD, Beauchamp RD, DuBois RN. Modulation of apoptosis and Bcl-2 expression by prostaglandin E2 in human colon cancer cells. Cancer Res 1998;58:362–6.
- Sheng H, Shao J, Washington MK, DuBois RN. Prostaglandin E2 increases growth and motility of colorectal carcinoma cells. J Biol Chem 2001;276:18075–81.
- Jones R, Adel-Alvarez LA, Alvarez OR, Broaddus R, Das S. Arachidonic acid and colorectal carcinogenesis. Mol Cell Biochem 2003;253:141–9.
- Six DA, Dennis EA. The expanding superfamily of phospholipase A2 enzymes: Classification and characterization. Biochim Biophys Acta 2000;1488:1–19.
- Cummings BS. Phospholipase A2 as target for anti-cancer drugs. Biochem Pharmacol 2007;74:949–59.
- Smith WL, Lagenbach R. Why there are two cyclooxygenase isozymes. J Clin Invest 2001;107:1491–5.
- Österström A, Dimberg J, Fransen K, Söderkvist P. Expression of cytosolic and group X secretory phospholipase A2 genes in human colorectal adenocarcinomas. Cancer Lett 2002;1982:175–82.
- Panel V, Boelle PY, Ayala-Sanmartin J, Jouniaux AM, Hamelin R, Masliah J, et al. Cytoplasmic phospholipase A2 expression in human colon adenocarcinoma is correlated with cyclooxygenase-2 expression and contributes to prostaglandin E2 production. Cancer Lett 2006;243:255–63.
- Yoo YS, Lim SC, Kim KJ. Prognostic significance of cytosolic phospholipase A2 expression in patients with colorectal cancer. J Korean Surg Soc 2011;80:397–403.
- Bumeida A, Bendardaf R, Hilska M, Laine J, Collan Y, Laato M, et al. PLA2 (group IIA phospholipase A2) as a prognostic determinant in stage II colorectal carcinoma. Ann Oncol 2009;20:1230–5.
- Murakami M, Masuda S, Kudo I. Arachidonate release and prostaglandin production by group IVC phospholipase A2 (cytosolic phospholipase A2γ). J Biochem 2003;372:695–702.
- Asai K, Hirabayashi T, Houjou T, Uozumit N, Taguchi R, Shimizu T. Human group IVC phospholipase A2 (cPLA2γ). Roles in the membrane remodeling and activation induced by oxidative stress. J Biol Chem 2003;278:8809–14.
- Yamashita A, Tanaka K, Kamata R, Kumazawa T, Suzuki N, Koga H, et al. Subcellular localization and lysophospholipase/transacylation activities of human group IVC phospholipase A2 (cPLA2 gamma). Biochim Biophys Acta 2009;1791:1011–22.
- Tian G, Wang X, Zhang F, Geng H, Hou W, Chen L, et al. Downregulation of cPLA2γ expression inhibits EGF-induced chemotaxis of human breast cancer cells through Akt pathway. Biochem Biophys Res Commun 2011;409:506–12.
- Goel A, Boland CR. Recent insights into the pathogenesis of colorectal cancer. Curr Opin Gastroenterol 2010;26:47–52.
- Mammano E, Belluco C, Bonafe M, Olivieri F, Mugianesi E, Barbi C, et al. Association of p53 polymorphisms and colorectal cancer: Modulation of risk and progression. Eur J Surg Oncol 2009;35:415–19.
- Theodoropoulos G, Papaconstantinou I, Felekouras E, Niketeas N, Karakitsos P, Panoussopoulos D, et al. Relation between common polymorphisms in genes related to inflammatory response and colorectal cancer. World J Gastroenterol 2006;12:5037–43.
- Tao R, Yu Y, Zhang X, Guo Y, Shi J, Zhang X, et al. Cytosolic PLA2 genes possibly contribute to the etiology of schizophrenia. Am J Med Genet B Neuropsychiatr Genet 2005;137B:56–8.
- Yu Y, Tao R, Shi J, Zhang X, Kou C, Guo Y, et al. A genetic study of two calcium-independent cytosolic PLA2 genes in schizophrenia. Prostaglandins Leukot Essent Fatty Acids 2005;73:351–4.
- Gertler R, Rosenberg R, Schuster T, Friess H. Defining a high-risk subgroup with colon cancer stages I and II for possible adjuvant therapy. Eur J Cancer 2009;45:2992–9.
- Sobrero A. Should adjuvant chemotherapy become standard treatment for patients with stage II colon cancer? For the proposal. Lancet Oncol 2006;7:515–16.
- O’Connor ES, Greenblatt DY, LoConte NK, Gangnon RE, Liou JI, Heise CP, et al. Adjuvant chemotherapy for stage II colon cancer with poor prognostic features. J Clin Oncol 2011;29:3381–8.
- Graziano F, Cascinu S. Prognostic molecular markers for planning adjuvant chemotherapy trials in Dukes’ B colorectal cancer patients: How much evidence is enough? Ann Oncol 2003;14:1026–38.
- Böckelman C, Engelman BE, Kaprio T, Hansen TF, Glimelius B. Risk of recurrence in patients with colon cancer stage II and III: A systematic review and meta-analysis of recent literature. Acta Oncol 2015:54:5–16.
- Glimelius B. Adjuvant chemotherapy for patients with rectal cancer – will the controversy be resolved? Acta Oncol 2015:54:433–6.
- Akiyoshi T, Kobunai T, Watanabe T. Recent approaches to identifying biomarkers for high-risk stage II colon cancer. Surg Today 2012;42:1037–45.
