Introduction
Hypoxia is a common feature of the microenvironment of solid tumors and the presence of hypoxic domains are associated with tumor aggressiveness [Citation1], insensitivity to radio- and chemotherapy, increased metastatic potential and poor survival[Citation2–5]. The adverse prognostic value of hypoxia has been the focus of intense research over the last decades and identification of hypoxia continues to be of major interest.
In a previous study [Citation6], we applied the 15-gene hypoxia signature by Toustrup et al. [Citation7] in patients with gastroesophageal cancer (GEC) in order to evaluate the prognostic impact of the signature. This signature comprises 15 hypoxia-induced and pH-independent genes and has shown to be prognostic in head and neck squamous cell carcinomas (HNSCC) treated without hypoxic modifications and to predict benefit from hypoxic modification of radiotherapy with nimorazole in HNSCC [Citation7]. In the previous study by Winther et al. [Citation6] (from here on referred to as the original study), the study population constituted of 51 patients with esophageal cancer (primarily ESCC) and 38 patients with esophago-gastric junction (EGJ) and gastric cancer (GC) [primarily adenocarcinoma (AC)]. Hierarchical cluster analysis of the 15 genes showed inter- and intra-group heterogeneity of gene expression between patients with ESCC and patients with EGJ/GC. This finding was indicative of hypoxia being more profound in ESCC compared with AC. In addition, gene expression was found to be a potential prognostic marker in patients with ESCC. However, the study population was small and the results did not reach statistical significance. The purpose of the present study was to reproduce these results in a new and larger, independent cohort of patients with GEC (from here on referred to as the present study).
Materials and methods
Patient population and treatment
In total, 131 patients diagnosed with histologically verified loco-regional GEC with available diagnostic, pre-therapeutic formalin-fixed paraffin-embedded (FFPE) tumor biopsies were included in the present study. Clinico-pathological data were retrospectively collected from patient medical records. All patients were treated with curative intend in the time period 2006–2013 from three independent clinical centers in Denmark: 1) Department of Oncology, Aarhus University Hospital; 2) Department of Oncology, Odense University Hospital; and 3) Rigshospitalet, Copenhagen University Hospital. The study was approved by The Central Denmark Region Committees on Health Research Ethics and the Danish Data Protection Agency and was conducted in accordance with the Helsinki Declaration.
Esophageal squamous cell carcinoma (present study)
Patient and tumor characteristics are summarized in . Overall, 89 patients were diagnosed with ESCC. Neoadjuvant chemoradiotherapy (concurrent 5-FU with/without cisplatin and 45–50 Gy) was given to 51 patients (57%) of whom 34 patients underwent surgery. Thirty-eight patients (43%) were allocated to definitive chemoradiotherapy (concurrent 5-FU with/without cisplatin and 50–60 Gy), however, three patients received radiotherapy (60 Gy) only due to age or co-morbidity. Among the patients allocated to definitive therapy, two patients underwent salvage surgery. Not all patients received full dose chemotherapy.
Table I. Patient and tumor characteristics.
Adenocarcinomas of the esophago-gastric junction and stomach (present study)
Forty-two patients were diagnosed with AC (. In total, 20 patients (48%) received perioperative chemotherapy (cisplatin, epirubicin and capecitabine) of whom 19 patients underwent surgery. Twenty patients (48%) were allocated to definitive chemoradiotherapy (concurrent 5-FU with/without cisplatin and 50–60 Gy), however, three patients received radiotherapy (60 Gy) only due to age or co-morbidity. Two patients (5%) were given neoadjuvant chemoradiotherapy (concurrent cisplatin, 5-FU and 45–50 Gy). One of the patients receiving neoadjuvant chemoradiotherapy underwent surgery. Not all patients received full dose chemotherapy.
RNA isolation, cDNA synthesis and gene expression analysis using qPCR
The procedure of gene expression analysis has previously been described by Winther et al. [Citation6]. Before analysis, presence of invasive carcinoma in the FFPEs was verified on hematoxylin and eosin stained sections. The mean fraction of tumor area (defined as the area of invasive carcinoma) was estimated to 63% (range 10–100).
Classification of hypoxic groups
The aim of the original study was to test the hypothesis that induction of the 15 hypoxia-responsive genes was associated with survival. The clinical impact of hypoxia gene expression was evaluated by ranking of gene expression and, subsequently, obtained by dividing patients into tertiles based on this ranking. In the present study, the potential prognostic value of the 15 genes in ESCC was sought reproduced by development of a classifier that enabled independent classification of the ESCC samples from the original and present study as either more or less hyoxic. Using the one-way ANOVA method described by Toustrup et al. [Citation8], ESCC patients in the original study were split into groups of more or less hypoxic tumors based on the greatest distance between the mean gene expression levels between the two groups. Based on the mean and variance of the gene expression in the two groups, the ESCC samples of the original and present study were independently classified as previously described by Toustrup et al. [Citation8].
Statistical analysis
Unsupervised hierarchical cluster analysis was performed in order to group patients with similar tumor gene expressions and, thus, elucidate the relationship between tumor histology and gene expression. Clustering analysis was performed with Gene Cluster version 3.0 using Pearson’s correlation and complete linkage and visualized using Treeview (Version 1.1.6r2).
The primary outcome was overall survival (OS), defined as time from date of histological diagnosis to death from any cause or last day of follow-up and the secondary outcome was disease-specific survival (DSS), defined as time from diagnosis to death from or with GEC or last day of follow-up. Survival curves were generated using the Kaplan-Meier method and survival data was expressed as hazard ratios (HR) using univariate Cox proportional hazards model. The proportional hazards assumption was tested using Schoenfeld residuals. In order to control for confounding, multivariate analysis by Cox proportional hazards model was performed with covariates of potential confounding (T- and N-stage). Statistical analysis was carried out using STATA, Version 12. All p-values are two-sided with a 5% level of significance. HR are presented with 95% confidence interval (CI).
Results
Outcome analysis
Esophageal squamous cell carcinomas (present study)
The median time of follow-up was 22 months (range 3–80) and the median OS was 24 months (95% CI 13–33). The overall one-year survival rate was 63% (95% CI 52–72) and the three-year OS rate 34% (95% CI 24–45). At evaluation time, 57 ESCC patients (64%) had died; the predominant cause of death was esophageal cancer (79%) whereas complications to treatment comprised 7% and other causes 14%.
No significant differences in age, sex, N- and M-stage, and OS were observed between ESCC patients in the present and original study. However, a significantly lower tumor size (p = 0.041) and T-stage (p = 0.003) were identified in the present cohort. In addition, treatment strategy differed between the two ESCC cohorts as definitive chemoradiotherapy was administered in ESCC patients in the present study.
Adenocarcinomas (present study)
In the AC group, patients were followed up for a median of 21 month (range 4–99). Median OS was 34 months (95% CI 15–54) with a one-year survival rate of 74% (95% CI 58–85) and a three-year survival of 44% (95% CI 27–59). Twenty-five patients (60%) died during the study period; 76% died from GEC, 8% from complications to treatment and 16% from other causes.
Comparing the AC groups of the present and original study, a significant difference between age (p = 0.021; higher in the present study) and N-stage (p = 0.012; lower in the present study) was observed. Again, treatment strategy differed between the cohorts of AC patients in that some patients from the present study received definitive chemoradiotherapy. No significant differences in sex, tumor size, T- and M-stage, and OS were identified.
Hypoxia-induced genes and tumor histopathology
The 15 hypoxia-induced and pH-independent genes of the 131 patients in the present study were subjected to unsupervised hierarchical clustering (). The analysis identified two patient clusters with tumors of more or less hypoxic genotypes. Induced gene expression indicated a more hypoxic genotype. Consistent with the original study [Citation6], inter-group heterogeneity between the ESCC and the AC group was observed. In addition, intra-group heterogeneity was identified in patients with ESCC, showing patients with more hypoxic genotypes and patients with less hypoxic genotypes. In contrast, patients with AC showed no intra-group variability and all patients were identified with tumors of less hypoxic genotypes.
Classification of hypoxic status of ESCC patients in the original study
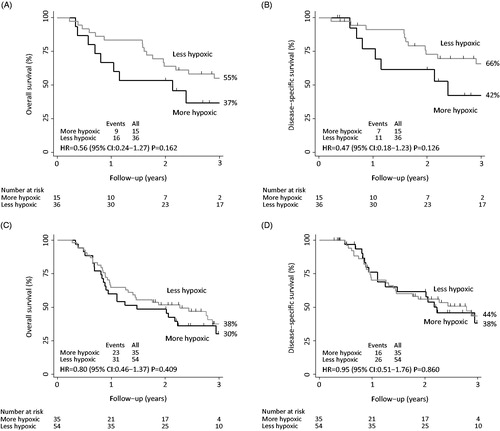
Using the one-way ANOVA method, the ESCC samples from the original study were independently classified into one group of 15 patients with more hypoxic genotypes and one group of 36 patients with less hypoxic genotypes. This classification was identical to the grouping of patients in the original study, except for one patient who was classified as more hypoxic in the original study and less hypoxic in the present. In line with the original results, outcome analysis for the newly generated groups showed a trend towards a better prognosis for patients with less hypoxic genotypes [OS: HR: 0.56 (95% CI 0.24–1.27); p = 0.162 and DSS: HR: 0.47 (95% CI 0.18–1.23); p = 0.126] (.
Figure 2. Kaplan-Meier estimates of (A) overall survival (original study), (B) disease-specific survival (original study), (C) overall survival (present study) and (D) disease-specific survival (present study) among patients with a more hypoxic genotype and a less hypoxic genotype based on the 15 hypoxia-induced and pH-independent genes.
Clinical impact of gene expression in ESCC patients in the present study
Based on the independent classification of the 89 ESCC samples in the present study, 35 patients were characterized with more hypoxic genotypes and 54 patients with less hypoxic genotypes. However, hypoxic status did not prove to be of prognostic significance in terms of OS [HR: 0.80 (95% CI 0.46–1.37); p = 0.409] or DSS [HR: 0.95 (95% CI 0.51–1.76); p = 0.860] in patients with ESCC (. Nor did a multivariate analysis adjusted for T- and N-stage [OS: HR: 0.87 (95% CI 0.49–1.53); p = 0.625 and DSS: HR: 0.97 (95% CI 0.51–1.86); p = 0.934]. When evaluating neoadjuvant and definitive chemoradiotherapy separately, no prognostic value of hypoxia was demonstrated for OS or DSS in either treatment group.
Discussion
This study sought to reproduce the clinical implications of gene expression of 15 hypoxia-inducible and pH-independent genes in a new and larger, independent cohort of 131 patients with GEC. The original study [Citation6] was indicative of hypoxia being more profound in patients with ESCC compared to AC, in addition to, gene expression being a potential prognostic marker in ESCC. Applying hierarchical cluster analyses, the present study confirmed the previous finding of inter- and intra-group heterogeneity of gene expression between ESCC and AC and, thus, confirmed that patients with ESCC held more hypoxic tumors and displayed greater heterogeneity compared to patients with AC. However, despite the larger sample size, the present study was not able to reproduce the previous indication of hypoxia gene expression serving prognostic value in ESCC patients.
The lack of ability to reproduce the prognostic impact may be explained by heterogeneity between the two ESCC patient populations. Thus, both data sets are collected retrospectively and not surprisingly differences in cT-stage exist between the two groups. It is acknowledged that differentiation between clinical assessments by either upper gastrointestinal endoscopy, CT or EUS into cT2 and cT3 is difficult. In addition, treatment regimens differs between the ESCC groups of the original and present study cohorts, as patients in the present study had received definitive chemoradiotherapy.
The prognostic potential of the 15-gene hypoxia signature in GEC was grounded in the properties of the signature. Thus, it has been shown to be prognostic in HNSCC [Citation7], and a subset of genes from the signature was overlapping with other prognostic hypoxia signatures [Citation9]. These overlapping genes included among others P4HA1, SLC2A1, BNIP3L, ALDOA and NDRG1, indicating that some genes are globally hypoxia-inducible regardless of cancer type or underlying approach.
In vitro studies have demonstrated that the 15 genes from the hypoxia signature by Toustrup et al. were hypoxia-inducible in sarcoma, prostate, colon, esophagus and EGJ cancer cell lines exposed to 0% oxygen [Citation10]. The present study showed upregulation of the 15 genes in ESCC but not in AC. These results indicate that hypoxia might be present to a lesser extend in this group of patients or that the 15-gene hypoxia signature might not be able to quantify hypoxia in patients with AC.
In conclusion, hypoxia is more profound in ESCC compared to AC. However, the study was not able to reproduce an association between hypoxia gene expression and survival, indicating that the 15-gene hypoxia signature may not have clinical prognostic implications in ESCC.
Acknowledgements
The authors thank Brita Singers Sørensen, Mogens Jøns Johannsen, Alice Baden and Nanna S. L. Hansen for excellent technical assistance. The study was financially supported by CIRRO – The Lundbeck Foundation Center for Interventional Research in Radiation Oncology and The Danish Council for Strategic Research, EC FP7 project METOXIA (Project no. 222741), The Danish Cancer Society, Karen A. Tolstrups Fund, The Family Spogaards Fund, The A.P. Moeller Foundation for the Advancement of Medical Science, Radiumstationens Forskningsfond, Max og Inger Wørzners Mindelegat, Frits, Georg og Marie Gluds legat, Fabrikant Willumsens Mindelegat and Dansk Kræftforsknings Fond.
Conflict of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Bertout JA, Patel SA, Simon MC. The impact of O2 availability on human cancer. Nat Rev Cancer 2008;8:967–75.
- Nordsmark M, Bentzen SM, Rudat V, Brizel D, Lartigau E, Stadler P, et al. Prognostic value of tumor oxygenation in 397 head and neck tumors after primary radiation therapy. An international multi-center study. Radiother Oncol 2005;77:18–24.
- Vaupel P, Mayer A. Hypoxia in cancer: significance and impact on clinical outcome. Cancer Metastasis Rev 2007;26:225–39.
- Harrison L, Blackwell K. Hypoxia and anemia: factors in decreased sensitivity to radiation therapy and chemotherapy? Oncologist 2004;9:31–40.
- Hockel M, Schlenger K, Aral B, Mitze M, Schaffer U, Vaupel P. Association between tumor hypoxia and malignant progression in advanced cancer of the uterine cervix. Cancer Res 1996;56:4509–15.
- Winther M, Alsner J, Tramm T, Nordsmark M. Hypoxia-regulated gene expression and prognosis in loco-regional gastroesophageal cancer. Acta Oncol 2013;52:1327–35.
- Toustrup K, Sørensen BS, Lassen P, Wiuf C, Alsner J, Overgaard J, et al. Gene expression classifier predicts for hypoxic modification of radiotherapy with nimorazole in squamous cell carcinomas of the head and neck. Radiother Oncol 2012;102:122–9.
- Toustrup K, Sørensen BS, Nordsmark M, Busk M, Wiuf C, Alsner J, et al. Development of a hypoxia gene expression classifier with predictive impact for hypoxic modification of radiotherapy in head and neck cancer. Cancer Res 2011;71:5923–31.
- Harris BH, Barberis A, West CM, Buffa FM. Gene Expression Signatures as Biomarkers of Tumour Hypoxia. Clin Oncol (R Coll Radiol) 2015;27:547–60.
- Sørensen BS, Knudsen A, Wittrup CF, Nielsen S, Aggerholm-Pedersen N, Busk M, et al. The usability of a 15-gene hypoxia classifier as a universal hypoxia profile in various cancer cell types. Radiother Oncol 2015; 116:346–351.

