Abstract
Background: In head and neck squamous cell carcinomas (HNSCC) hypoxic radioresistance can be reduced by use of the hypoxic modifier nimorazole, as shown in the DAHANCA 5 trial. Recently, a 15-gene hypoxia classifier has shown predictive impact for the effect of nimorazole by identifying ‘more’ and ‘less’ hypoxic tumors in the DAHANCA 5 cohort. A prospective multicentre EORTC-1219 study is initiated, where nimorazole and prospective use of the classifier as a predictor is tested in relation to the most recent accelerated chemoradiotherapy treatment. Validation of the gene expression classification procedures is described here.
Material and methods: Formalin-fixed paraffin-embedded (FFPE) tumor material from three recent HNSCC cohorts [DAHANCA 18 (n = 96), 24 (n = 40), and IAEA Hypo (n = 55)] was used to establish and validate procedures for prospective classification of patients. Repeatability was tested for the different steps in the gene expression analysis, and reproducibility was tested with xenograft tumors (FaDuDD, UTSCC33), where gene expression in complementary sections was compared after fixation and embedding locally and at international institutions, respectively. Intra-tumor heterogeneity was addressed by classifying biopsy samples from HNSCC tumors, where 2–4 biopsies from each tumor was accessible.
Results: Procedures were successfully established for individual classification of HNSCC patients in retrospective and prospective cohorts. Measurements of gene expression levels were reproducible between different international institutions.
Conclusion: Technical validation of the 15-gene hypoxia classifier demonstrated that it is suitable for implementation in prospective clinical trials.
In head and neck squamous cell carcinomas (HNSCC) hypoxic radioresistance can be reduced by concomitant use of the hypoxic modifier nimorazole. This was shown more than 20 years ago by the Danish Head and Neck Cancer Group in the DAHANCA 5 trial [Citation1]. Despite the results from this trial and a subsequent meta-analysis [Citation2], hypoxic modification with nimorazole is only used routinely in Denmark and in patients participating in DAHANCA trials [Citation3]. Benefit form hypoxic modification is expected to be most pronounced in patients with more hypoxic tumors. However, there are no validated imaging or molecular biomarkers approved by the US Food and Drug Administration (FDA) or European Medicines Agency (EMA) that can estimate the level of hypoxia and predict benefit from hypoxic modifications like nimorazole.
A 15-gene hypoxia classifier has been developed, which is able to identify ‘more hypoxic’ and ‘less hypoxic’ tumors in the DAHANCA 5 trial randomizing patients to conventional radiotherapy with either placebo or nimorazole [Citation4]. The classifier was developed from in vitro data on HNSCC cell lines [Citation5], tested in xenograft models of these cell lines and finally defined in a training set of HNSCC patients with hypoxia-evaluated tumors (by oxygen electrode measurements) [Citation4,Citation6]. The 15-gene hypoxia classifier was developed to classify patients individually, and in the DAHANCA 5 trial, the classifier demonstrated both prognostic and predictive value. The prognostic value was seen in the placebo group with significantly worse outcome in patients with more hypoxic tumors. The classifier is also predictive, as the benefit of nimorazole was only observed in the one third of patients with more hypoxic tumors [Citation4]. The frequency of ‘more’ and ‘less’ hypoxic tumors, when analyzed with the 15-gene hypoxia classifier, was independent of HPV/p16-status [Citation7,Citation8]. However, in the DAHANCA 5 trial, nimorazole was shown to only improve outcome in patients with HPV/p16-negative tumors, and not in HPV/p16-positive [Citation7,Citation9]. In Denmark, the prevalence of HPV/p16-positive HNSCC tumors has increased dramatically in oropharyngeal carcinomas since the DAHANCA 5 trial was performed in the late 1980s [Citation10,Citation11]. With the current Danish treatment regime of accelerated radiotherapy with concomitant chemotherapy and nimorazole, five-year loco-regional control rates of more than 90% are now observed in HPV/p16-positive oropharyngeal carcinomas [Citation12]. At present, it is not known to what extent nimorazole improves outcome in HNSCC patients receiving modern treatment regimens [Citation13,Citation14], whether the 15-gene hypoxia classifier has predictive value, and whether the effect of nimorazole (and predictive value of the classifier) is limited to patients with HPV/p16-negative tumors. Recent studies did confirm that the 15-gene hypoxia classifier has prognostic value in HNSCC patients with HPV/p16-negative tumors receiving modern treatment regimens without nimorazole [Citation8] and in a cohort of HNSCC patients from The Cancer Genome Atlas [Citation15].
The effect of nimorazole has recently been tested in combination with accelerated radiotherapy (without concomitant chemotherapy) in the IAEA Hypo trial [Citation16]. The trial was activated in centers in Egypt, Pakistan, Estonia, and Slovenia, but was closed prematurely due to the poor recruitment rate. Overall, nimorazole was associated with a non-significant improvement in outcome. A subset of patients was analyzed retrospectively for the 15-gene hypoxia classifier. Patients with more hypoxic tumors, if treated with radiotherapy alone, had a significantly higher risk of loco-regional tumor failure as compared to the rest of the patients.
Currently, the effect of nimorazole is being investigated in two randomized trials. The UK NIMRAD trial ([Citation17], ClinicalTrials.gov Identifier: NCT01950689) includes HNSCC patients not suitable for synchronous chemotherapy and receiving accelerated radiotherapy alone ± nimorazole. A retrospective analysis is planned for the predictive value of a 25/26-gene hypoxia classifier [Citation18,Citation19] and the 15-gene hypoxia classifier [Citation4,Citation7]. The international multicentre EORTC-1219 study includes HPV/p16-negative HNSCC patients receiving the most recent accelerated chemoradiotherapy treatment regime ± nimorazole (ClinicalTrials.gov Identifier: NCT01880359). This trial has two independent primary objectives, to test the overall effect of nimorazole and to test the predictive value of the 15-gene hypoxia classifier. The analysis of the 15-gene hypoxia classifier is therefore performed prospectively at a central laboratory and the result of the test is included in the stratification procedure. The test is performed on routine formalin-fixed paraffin-embedded (FFPE) tumor biopsies, and this paper describes the technical validation studies of the test in relation to measurement and normalization of gene expression levels, reproducibility, repeatability, and tumor heterogeneity.
Material
FFPE tumor material was obtained from 514 patients from five head and neck cohorts ( for patient characteristics). The cohorts include DAHANCA 5 (1986–1990, n = 323) [Citation1], DAHANCA 18 (2007–2010, n = 96) [Citation12], DAHANCA 24 (2009–2011, n = 40) [Citation20], and IAEA HypoX (2012–2014, n = 55, patients from Egypt, Pakistan, Estonia, and Slovenia) [Citation16]. Four patients were included in both DAHANCA 18 and 24, leaving 510 unique patients. Presence of invasive carcinoma was verified on hematoxylin/eosin (HE) sections.
Table 1. Patient and tumor characteristics in cohorts.
Tumor xenografts (FaDuDD and UTSCC33) were grown as subcutaneous flank tumors, established by injection of cells into NMRI-nu/nu female mice. Both cell lines were obtained from Dr. Michael Baumann, University of Technology, Dresden, Germany.
Methods
Xenograft tumors (150–1000 mm3) were divided in two immediately after excision and snap frozen. For internal reproducibility studies at the central laboratory [Department of Experimental Clinical Oncology, Aarhus University Hospital, Denmark (ECO)], both halves of xenograft tumors were formalin-fixed and paraffin-embedded. External reproducibility was tested by keeping and paraffin-embedding one half of a xenograft tumors at ECO, sectioning these tumors from the cut side, and sending the other halves as frozen samples to a local pathology department (Department of Pathology, Aarhus University Hospital, Denmark) and to seven different external international pathology centers involved in the EORTC-1219 study. The external centers performed their usual fixation and embedding procedure, sectioned the tumors from the cut side and returned the test-tumor sections to ECO for gene expression quantification. Subsequently, gene expression from tumors sections preserved at external centers was compared to their complementary sections kept at ECO.
For both xenograft tumors and patient biopsies, FFPE samples were sectioned, RNA extracted, and gene expression levels were determined by qPCR as described previously [Citation4]. For xenograft tumors, gene expression was measured as delta Ct values (ΔCt). For patient biopsies, a further normalization step was introduced in order to compensate for technical developments, allowing measurements in recent samples to be compared to the original values from the DAHANCA 5 trial (see Supplementary Document for further details).
Correlations between gene expression levels in xenograft tumors were evaluated by Lin’s Concordance correlation coefficient, ρC, and a Bland-Altman plot with 95% limits of agreement (LOA) for each gene in the 15-gene classifier. Two-class k-means clustering was performed using Euclidian distance. Pearson’s χ2-test was used for correlation between p16 and classification. Statistical analyses were performed using Stata 14.
Results
Repeatability
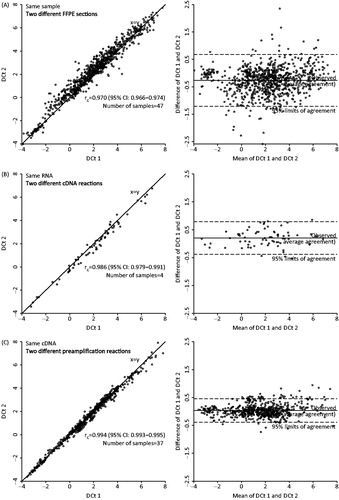
The repeatability and the technical variability was established from all steps in qPCR procedures by independent replicate analysis of the same samples. Repeatability of RNA extraction was verified in 47 samples from the DAHANCA 18 cohort, where RNA was extracted from two different sections. The RNA extraction and all downstream procedures were carried out in independent procedures. Comparison of the ΔCt levels from the two sections show a high grade of comparability (), with ρC = 0.970 and 95% LOA of −1.21 and 0.68. The repeatability of the qPCR procedure was verified from both RNA (four samples from the DAHANCA 18 cohort, ), and cDNA (37 samples from the DAHANCA 18 cohort, ). For independent analysis on same RNA, ρC = 0.986 and 95% LOA (−0.38–0.79). For independent analysis on same cDNA, ρC = 0.994 and 95% LOA (−0.39–0.46).
Figure 1. Repeatability and technical variability in the various steps in the qPCR procedures by independent replicate analysis of the same samples and plotted as correlations of delta Ct levels and as mean versus difference (Bland-Altman plot). Includes the 15 hypoxia-responsive genes and the three reference genes. Correlation of delta Ct levels in two different sections from the same sample (A). Delta Ct values from the same sample, repeated from RNA level (B). Delta Ct values from the same sample, repeated from cDNA level (C).
Reproducibility
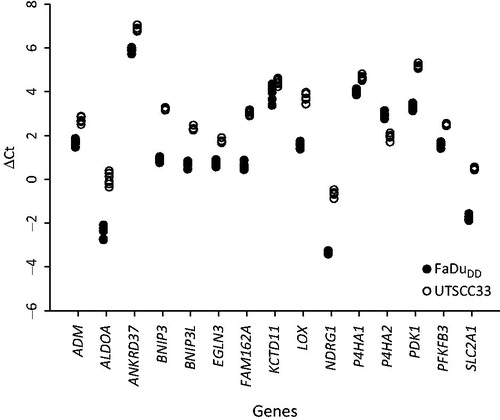
Tumor xenografts (FaDuDD and UTSCC33) were used to test for the influence of variation in fixation procedure on gene expression levels and reproducibility of the procedure. First, six sections from each of two tumor samples (one FaDuDD and one UTSCC33) were analyzed (). The differences in ΔCt between the minimum and maximum values for each gene in the six sections was on average 0.40 for FaDuDD (range 0.16–0.98) and 0.32 for UTSCC33 (range 0.11–0.74).
Figure 2. Delta Ct values for the 15 hypoxia-responsive genes from six sections from the same xenograft tumor samples.
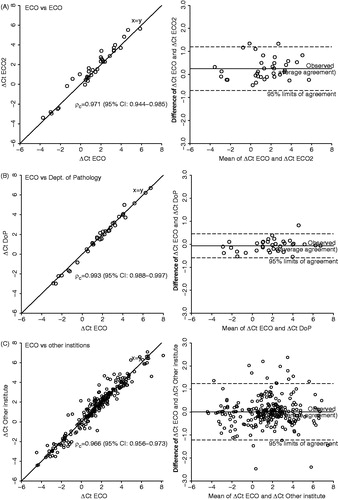
Next, sets of tumor samples (each set containing one FaDuDD and one UTSCC33) were cut in half, fixed and embedded individually (one half always at the central laboratory, ECO), and two sections from the cut side of each half of the tumors were compared (). When the other halves were also fixed and embedded at ECO, the ρC for the agreement of ΔCt values was 0.97 and the 95% LOA was -0.69–1.20 (). When the other halves were frozen and then fixed, embedded, and cut at the local pathology department (DoP), ρC was 0.99 and LOA was -0.58–0.46 (). Sets of tumor samples were also send frozen to seven different centers in Europe and fixed, embedded, and cut using standard procedures at each center. shows that for the combined data for all comparisons between sections from the tumor halves kept at ECO and the corresponding halves at the different institutions, ρC was 0.97 and LOA was −1.22–1.22. For five of the institutions, ρC>0.96, and for the remaining two ρC was 0.94 (95% CI 0.88–0.97) and 0.90 (95% CI 0.82–0.95).
Figure 3. Reproducibility of the procedure tested by correlation of delta Ct levels of the 15 hypoxia-responsive genes and the three reference genes from two sections from each half of tumor pairs (each pair consisting of one FaDuDD and one UTSCC33) with one half of the tumors always fixed, embedded and cut at the central laboratory (ECO). Paired delta Ct levels are plotted against each other and as mean versus difference (Bland-Altman plot). Correlation of delta Ct levels of sections with the second halves fixed, embedded, and cut at ECO (A), at the local pathology department at Aarhus University Hospital (DoP) (B), and at seven different institutions in Europe (C).
Classification
In order to classify patients prospectively using the same classifier as described in the original study on the DAHANCA 5 cohort [Citation4], expression values for the recent cohorts were normalized to a cohort consisting of 108 patients from DAHANCA 18 or 24 with supraglottic laryngeal, hypopharyngeal, or oropharyngeal cancer (Supplementary Table 1, available online at http://www.informahealthcare.com).
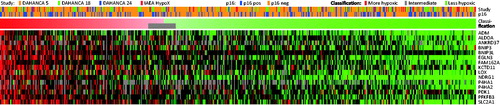
shows the classification results for 510 patients from DAHANCA 5, 18, 24 and IEAE HypoX. The data for DAHANCA 5 are from the original study [Citation4], and the remaining patients are classified as described in the Supplementary Document (available online at http://www.informahealthcare.com). As in the original DAHANCA 5 study, 35% of the patients were classified as more hypoxic patients in the four recent cohorts (range 29–40%). Patients in are ranked from the most to the least hypoxic, and there is no apparent correlation between the rank of the patients and the cohorts. There was no difference in the frequency of patients classified as more hypoxic in p16 negative and p16 positive tumors (34% vs. 38%, p = 0.28). Patients can either all be classified as ‘more’ or ‘less’ hypoxic, or a subset of patients can be categorized as ‘intermediate’ (6%) (see Supplementary Document for details). Both means of classification are shown in .
Figure 4. Classification of 510 independent samples from DAHANCA 5, 18, 24, and IAEA HypoX. Gene expression levels are shown as heatmap (each gene centered on the mean of the values characterizing ‘more’ and ‘less’ hypoxic tumors).
Two-class k-means clustering was performed on the 510 patients in , resulting in a ‘high expression’ and a ‘low expression’ group. Due to missing values, k-means clustering could not be performed on 186 patients. Of the remaining 324 patients, all 109 classified as ‘more hypoxic’ clustered in the ‘high expression’ group. Of the 215 classified as ‘less hypoxic’, 143 (67%) clustered in the ‘low expression’ group and 72 (33%) clustered in the ‘high expression’ group. Thus, classification based on the original classifier identified 34% of the patients as ‘more hypoxic’ and k-means clustering identified 56% in the ‘high expression’ group, which is interpreted as the hypoxic patients.
Heterogeneity within classifier genes
Classifications were performed on each of the genes included in the classifier and the results were matched with the overall classification (). On average, single gene classifications matched the overall classification in 72% of the cases for the four recent cohorts (range 60–80%). Due to the age of the FFPE blocks, the DAHANCA 5 cohort contained a number of missing values, but displayed similar results with single gene classifications matching the overall classification in 72% of the cases (range 62–80%).
Table 2. Heterogeneity within classifier genes.
Intra-tumor heterogeneity
Intra-tumor heterogeneity was addressed in 20 tumors from the DAHANCA 18 trial, where 2–4 different biopsies from each tumor were accessible (n = 53). Fraction of tumor area, defined as area of invasive carcinoma as a percentage of the total biopsy area, was semi-quantitatively estimated from HE sections by light microscopy, and each biopsy was classified. In 14 of the 20 tumors there was full concordance between classification of all samples from the same tumor (). In four tumors, the biopsy containing the relatively highest proportion of tumor tissue was classified as more hypoxic. In the remaining two tumors (shaded area in ), the biopsy with the highest tumor tissue content was classified as ‘less’ hypoxic, but another biopsy with slightly less relative tumor tissue content was classified as ‘more’ hypoxic.
Figure 5. Intra-tumor heterogeneity. Classification of 20 patients with multiple biopsies, with shaded area indicating two patients where the biopsy with the highest tumor content did not reveal hypoxia, but another biopsy did (A). Classification of 129 samples from 96 patients plotted against tumor content in biopsy (B).
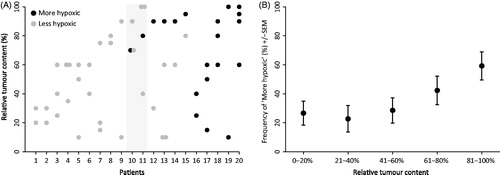
To examine a possible limit of detection linked to the relative tumor content of a biopsy, 129 samples from 96 patients in the DAHANCA 18 cohort were compared (). There does not seem to be a lower level where samples cannot be classified as’ more hypoxic’, and in biopsies below 60% tumor tissue content, the frequency of ‘more hypoxic’ tumors is stable. In biopsies with more than 60% tumor tissue content, there is a tendency for a higher frequency of ‘more hypoxic’ tumors.
Discussion
Several hypoxia gene expression signatures have been developed and have shown promising prognostic impact when tested retrospectively in historical clinical cohorts, as reviewed in [Citation21,Citation22]. Here, we describe the technical validation for the prospective use of a 15-gene hypoxia classifier to predict benefit from hypoxia modification.
The validation included measurements of repeatability, reproducibility, classification, and intra-tumor heterogeneity. Repeatability was tested from three different steps in the measurement of gene expression by qPCR, by comparing RNA extracted from different sections from the same FFPE block, and by independent measurements starting from the same RNA and the same cDNA. Overall, repeatability was high and, as expected, increased during each of the three steps with measurements from the same cDNA having the highest repeatability. Thus, in accordance with a previous study on the same analytical procedures [Citation23], technical variability is considered negligible.
Reproducibility is an important issue when a gene expression classifier is applied to sections from routine FFPE biopsies from different international institutions. Reproducibility was tested using xenograft tumor models where individual tumors were cut in halves. One half was fixed, embedded, at cut at the central laboratory. The other half was processed independently at the central laboratory, or frozen and send to different international institutions participating in the EORTC 1219 trial (ClinicalTrials.gov Identifier: NCT01880359). The institutions fixed and embedded the samples according to routine guidelines, and sectioned the blocks from the cut side and returned the sections to the central laboratory where gene expression levels were compared with sections from the corresponding halves of the tumors. Differences in routine fixation procedures give rise to some variability in gene expression levels, but no systematic differences were observed. Compared to the variability within a tumor block sectioned six times, and the variability when two halves were processed individually at the central laboratory, it is concluded that it is acceptable to use the local routine fixation procedures at different international institutions.
For prospective studies where patients are classified prior to treatment, or prior to stratification in a clinical trial, classification needs to be done individually. The original classifier [Citation4] was developed in order to provide individual classification and is based on defined expression values characteristic of ‘more’ and ‘less’ hypoxic tumors. Here, we have described how absolute expression levels of recent samples can be normalized, allowing independent classification of patients where gene expression is measured on a different platform and/or a different scale as compared to the original cohort. This robustness of the procedure was demonstrated by successfully applying it to patients from Denmark, Egypt, Pakistan, Estonia, and Slovenia.
Recently, the prognostic potential of different hypoxia gene classifiers has been compared in HNSCC cohorts [Citation8,Citation15]. These studies have used a different method of classification (k-means clustering). The advantage of k-means clustering is that it can group the same cohort of patients based only on different lists of gene names (classifiers) and the expression values of these genes. However, when k-means clustering is applied to the patients included here, the frequency of ‘more hypoxic’ patients change from 35% based on the original classification procedure to 56% based on k-means clustering. Thus, even though the same genes are analyzed in the same cohort of patients, the prognostic potential will be significantly different when applying different classification procedures.
In the DAHANCA 5 study, the frequencies of ‘more’ and ‘less’ hypoxic tumors were independent of HPV/p16-status [Citation7,Citation8]. The same was found in a recent study from the German Cancer Consortium Radiation Oncology Group [Citation8] and for the DAHANCA 18, 24, and IAEA Hypo X cohorts reported here. Furthermore, in vitro data have demonstrated that there is no difference in hypoxia induced gene expression of the 15 genes in the classifier when comparing HPV positive and HPV negative cell lines [Citation24]. Thus, there is no correlation between the 15-gene classifier and HPV/p16-status. This is in contrast to the 25/26-gene signature, where there is a lower frequency of tumors with high expression levels of hypoxia-responsive genes in HPV/p16 positive patients [Citation8,Citation19].
To improve the chances that a gene expression classifier can be applied to routine clinical setting across different institutions in different countries, the methods and procedures have to be relatively simple and robust. With increasing focus on shortening waiting time, especially for HNSCC patients treated with radiotherapy [Citation25,Citation26], time-to-result can also be a major issue. The 15-gene classifier has been developed to work on a single section of a routine diagnostic FFPE blocks, and with the current setup at the central laboratory in the EORTC 1219 trial it takes two days from starting with a cut section to classification is performed. Robustness is further ensured by using up to 15 genes in the signature. Although they are all hypoxia-responsive genes, individual variations do occur with different genes in different patients. Thus, on average single gene based classifications are found to match the overall classification of the samples in 72% of the cases. Not all 15 genes have to be measured in order to perform the classification, as the procedure is designed to handle missing values. In the original study on the DAHANCA 5 cohort missing values was an issue, due to the quality of the RNA in the older paraffin sections, but with recent cohorts and in prospective analysis missing values are less common.
Intra-tumor heterogeneity is a major issue for biomarker studies based on single tumor biopsies, and tumors are known to be heterogeneous in regards to the extent of hypoxia [Citation27]. We tested intra-tumor heterogeneity by classifying multiple biopsies with different tumor tissue content from 20 individual patients. In 14 patients, all biopsies from the same patient were classified the same. In four patients the biopsy with the highest tumor tissue content classified as ‘more’ hypoxic, but other biopsies with low tumor content were classified as ‘less’ hypoxic. In two patients the biopsy with the highest tumor content classified as ‘less’ hypoxic, but another biopsy with slightly less tumor content was classified as ‘more’ hypoxic. We also attempted to identify a potential cut-point for minimum tumor tissue content. However, below 60% tumor content, the frequencies of ‘more’ hypoxic tumors appeared stable. Above 60% tumor content, the frequencies of ‘more’ hypoxic tumors appeared to increase. In order keep the procedures relatively simple and robust, while still keeping the number of missed ‘more’ hypoxic tumors to a minimum, the current recommendation to the pathologists cutting the FFPE blocks is therefore if multiple biopsies are available, to select the optimal biopsy taking the relative content of invasive carcinoma, minimal necrosis and the total size of the tumor biopsy into account.
In conclusion, this technical validation of the 15-gene hypoxia classifier has demonstrated that it is suitable for further implementation and evaluation of prognostic impact and predictive value for the use of hypoxia-modifying therapies.
Supplementary_Document.docx
Download MS Word (29.3 KB)Supplementary_Document.pdf
Download PDF (469.3 KB)Acknowledgments
The authors would like to thank Mogens Johansen, Alice Baden and Nanna Nielsen for technical assistance. Cell lines were obtained with courtesy of Dr. Michael Baumann. Financial support was received from the Danish Cancer Society, CIRRO – The Lundbeck Foundation Center for Interventional Research in Radiation Oncology and The Danish Council for Strategic Research.
Disclosure statement
K. Toustrup, B. S. Sørensen, J. Overgaard and J. Alsner are listed as co-inventors on a provisional patent application on a method for determining clinically relevant hypoxia in cancer (WO/2012/146259) that is owned by Aarhus University, Aarhus, Denmark, and the part concerning prediction of benefit from Nimorazole is licensed to Azanta Denmark A/S, Hellerup, Denmark. None of the authors have gained financially from the patent and no commercial funding has been provided in the generation of data for this paper.
References
- Overgaard J, Hansen HS, Overgaard M, Bastholt L, Berthelsen A, Specht L, et al. A randomized double-blind phase III study of nimorazole as a hypoxic radiosensitizer of primary radiotherapy in supraglottic larynx and pharynx carcinoma. Results of the Danish Head and Neck Cancer Study (DAHANCA) Protocol 5-85. Radiother Oncol 1998;46:135–46.
- Overgaard J. Hypoxic modification of radiotherapy in squamous cell carcinoma of the head and neck-a systematic review and meta-analysis. Radiother Oncol 2011;100:22–32.
- Overgaard J. Hypoxic radiosensitization: Adored and ignored. J Clin Oncol 2007;25:4066–74.
- Toustrup K, Sørensen BS, Nordsmark M, Busk M, Wiuf C, Alsner J, et al. Development of a hypoxia gene expression classifier with predictive impact for hypoxic modification of radiotherapy in head and neck cancer. Cancer Res 2011;71:5923–31.
- Sørensen BS, Toustrup K, Horsman MR, Overgaard J, Alsner J. Identifying pH independent hypoxia induced genes in human squamous cell carcinomas in vitro. Acta Oncol 2010;49:895–905.
- Nordsmark M, Overgaard J. A confirmatory prognostic study on oxygenation status and loco-regional control in advanced head and neck squamous cell carcinoma treated by radiation therapy. Radiother Oncol 2000;57:39–43.
- Toustrup K, Sørensen BS, Lassen P, Wiuf C, Alsner J, Overgaard J. Gene expression classifier predicts for hypoxic modification of radiotherapy with nimorazole in squamous cell carcinomas of the head and neck. Radiother Oncol 2012;102:122–9.
- Linge A, Lock S, Gudziol V, Nowak A, Lohaus F, von NC, et al. Low cancer stem cell marker expression and low hypoxia identify good prognosis subgroups in HPV(-) HNSCC after postoperative radiochemotherapy: A Multicenter Study of the DKTK-ROG. Clin Cancer Res 2016. [Epub ahead of print]. DOI: 10.1158/1078-0432.CCR-15-1990.
- Lassen P, Eriksen JG, Hamilton-Dutoit S, Tramm T, Alsner J, Overgaard J. HPV-associated p16-expression and response to hypoxic modification of radiotherapy in head and neck cancer. Radiother Oncol 2010;94:30–5.
- Lassen P. The role of Human papillomavirus in head and neck cancer and the impact on radiotherapy outcome. Radiother Oncol 2010;95:371–80.
- Lassen P, Primdahl H, Johansen J, Kristensen CA, Andersen E, Andersen LJ, et al. Impact of HPV-associated p16-expression on radiotherapy outcome in advanced oropharynx and non-oropharynx cancer. Radiother Oncol 2014;113:310–16.
- Bentzen J, Toustrup K, Eriksen JG, Primdahl H, Andersen LJ, Overgaard J. Locally advanced head and neck cancer treated with accelerated radiotherapy, the hypoxic modifier nimorazole and weekly cisplatin. Results from the DAHANCA 18 phase II study. Acta Oncol 2015;54:1001–7.
- Baujat B, Bourhis J, Blanchard P, Overgaard J, Ang KK, Saunders M, et al. Hyperfractionated or accelerated radiotherapy for head and neck cancer. Cochrane Database Syst Rev 2010;12:CD002026.
- Blanchard P, Baujat B, Holostenco V, Bourredjem A, Baey C, Bourhis J, et al. Meta-analysis of chemotherapy in head and neck cancer (MACH-NC): A comprehensive analysis by tumour site. Radiother Oncol 2011;100:33–40.
- Tawk B, Schwager C, Deffaa O, Dyckhoff G, Warta R, Linge A, et al. Comparative analysis of transcriptomics based hypoxia signatures in in head- and neck squamous cell carcinoma. Radiother Oncol 2016;118:350–8.
- Hassan Metwally MA, Ali R, Kuddu M, Shouman T, Strojan P, Iqbal K, et al. IAEA-HypoX. A randomized multicenter study of the hypoxic radiosensitizer nimorazole concomitant with accelerated radiotherapy in head and neck squamous cell carcinoma. Radiother Oncol 2015;116:15–20.
- Thomson D, Yang H, Baines H, Miles E, Bolton S, West C, et al. NIMRAD – a phase III trial to investigate the use of nimorazole hypoxia modification with intensity-modulated radiotherapy in head and neck cancer. Clin Oncol (R Coll Radiol) 2014;26:344–7.
- Eustace A, Mani N, Span PN, Irlam JJ, Taylor J, Betts GN, et al. A 26-gene hypoxia signature predicts benefit from hypoxia-modifying therapy in laryngeal cancer but not bladder cancer. Clin Cancer Res 2013;19:4879–88.
- Betts GN, Eustace A, Patiar S, Valentine HR, Irlam J, Ramachandran A, et al. Prospective technical validation and assessment of intra-tumour heterogeneity of a low density array hypoxia gene profile in head and neck squamous cell carcinoma. Eur J Cancer 2013;49:156–65.
- Mortensen LS, Johansen J, Kallehauge J, Primdahl H, Busk M, Lassen P, et al. FAZA PET/CT hypoxia imaging in patients with squamous cell carcinoma of the head and neck treated with radiotherapy: results from the DAHANCA 24 trial. Radiother Oncol 2012;105:14–20.
- Toustrup K, Sørensen BS, Alsner J, Overgaard J. Hypoxia gene expression signatures as prognostic and predictive markers in head and neck radiotherapy. Semin Radiat Oncol 2012;22:119–27.
- Harris BH, Barberis A, West CM, Buffa FM. Gene expression signatures as biomarkers of tumour hypoxia. Clin Oncol (R Coll Radiol) 2015;27:547–60.
- Søes S, Sørensen BS, Alsner J, Overgaard J, Hager H, Hansen LL, et al. Identification of accurate reference genes for RT-qPCR analysis of formalin-fixed paraffin-embedded tissue from primary non-small cell lung cancers and brain and lymph node metastases. Lung Cancer 2013;81:180–6.
- Sørensen BS, Busk M, Olthof N, Speel EJ, Horsman MR, Alsner J, et al. Radiosensitivity and effect of hypoxia in HPV positive head and neck cancer cells. Radiother Oncol 2013;108:500–5.
- Jensen AR, Nellemann HM, Overgaard J. Tumor progression in waiting time for radiotherapy in head and neck cancer. Radiother Oncol 2007;84:5–10.
- van Harten MC, Hoebers FJ, Kross KW, van Werkhoven ED, van den Brekel MW, van Dijk BA. Determinants of treatment waiting times for head and neck cancer in the Netherlands and their relation to survival. Oral Oncol 2015;51:272–8.
- Horsman MR, Mortensen LS, Petersen JB, Busk M, Overgaard J. Imaging hypoxia to improve radiotherapy outcome. Nat Rev Clin Oncol 2012;9:674–87.
