Abstract
Interindividual variation of blood pressure (BP) responses to antihypertensive drugs is extensive. Several clinical, laboratory, and genetic predictors of BP responses to blood pressure-lowering agents have been suggested. We describe here the principal findings from the GENRES Study which is primarily a pharmacogenetic study of antihypertensive drug responses but also includes analysis of certain clinical and laboratory predictors. In this placebo-controlled, double-blinded, and randomized study, more than 200 male subjects with essential hypertension were treated with four antihypertensive drug monotherapies (amlodipine, bisoprolol, hydrochlorothiazide, and losartan) in a cross-over fashion, resulting in more than 800 treatment periods. Generally, placebo BP level was the best predictor of BP responses. In addition, higher baseline plasma renin activity predicted better BP response to losartan and bisoprolol, and weaker response to hydrochlorothiazide. A number of candidate gene polymorphisms analysed so far have given negative results in relation to BP responses, with the exception of an STK39 variant associating with losartan responsiveness. In future, genome-wide association studies on antihypertensive pharmacogenetics may identify novel pathways of BP regulation and provide new tools for both basic research and clinical use.
| Abbreviations | ||
| ABP | = | ambulatory blood pressure |
| BP | = | blood pressure |
| DBP | = | diastolic blood pressure |
| GWAS | = | genome-wide association studies |
| OBP | = | office blood pressure |
| PRA | = | plasma renin activity |
| SBP | = | systolic blood pressure |
Key messages
Several predictors of antihypertensive responses have been identified.
No genetic polymorphisms can currently be used in clinical practice to predict blood pressure responses in essential hypertension.
Large-scale genomic approaches may identify novel pathways of blood pressure regulation.
Introduction
Although hypertensive patients as a group respond to most antihypertensive drug treatments with a reasonably predictable blood pressure (BP) decrease, the individual responses are highly variable. In occasional cases, BP responses in essential hypertension may compare to those seen in cases with monogenic forms of hypertension where mutation-directed tailored treatments are initiated, while in other cases no response or even increased BP levels are observed, whether encountered in clinical practice or in controlled studies (Citation1,Citation2).
The search for predictors of antihypertensive drug responses has two motives. First, clinical treatment of hypertension requires intensification, since few treated patients reach target BP levels (Citation3,Citation4). Although most patients in clinical trials appear to need combination therapy to reach desirable BP levels when treatment protocols are fixed, rotational studies have shown that monotherapy may be effective in the majority of patients if the proper drug can be found (Citation5). Availability of robust predictors of BP response could facilitate success in treatment. Second, BP is a tricky phenotype to assess. In addition to the difficulties in obtaining reliable BP measurements, multiple known and unknown genetic and lifetime environmental factors and physiological pathways affect BP levels and underlying regulatory mechanisms. Genome-wide association studies (GWAS) on population BP levels have given a number of positive results, but only with small effect sizes after expansion of subject numbers to several tens of thousands, possibly because these studies were mostly based on single time point BP measurements with high intraindividual variability. On the other hand, using BP response to a specific pharmacological therapy as the phenotype may provide an alternative way to identify gene loci influencing BP levels, as use of BP levels prior to and during drug treatment may partially control for interindividual contribution of environmental factors.
In this mini-review, we present data on the variability and the predictors of BP responses to antihypertensive treatment from the GENRES Study (Citation2).
Methodological aspects of the GENRES Study
GENRES is a double-blind, placebo-controlled, randomized, and cross-over study with four 4-week monotherapies from the major antihypertensive drug groups (losartan, bisoprolol, amlodipine, and hydrochlorothiazide). Both 24-h ambulatory (ABP) and office (OBP) blood pressure measurements were carried out. More than 200 middle-aged (35–59 years) Finnish men completed the study, resulting in more than 800 drug treatment periods. There were 4-week placebo periods before and after each treatment period. Consequently, the placebo BP level estimates should be very accurate, since they are derived from four different placebo periods. Estimation of the number of subjects required was based on a priori power calculations (Citation2). For specific genetic polymorphisms, post-hoc power calculations may be performed. For example, assuming genotype group numbers of 180 (wild-type homozygotes) and 25 (variant allele heterozygotes), alpha level of 0.05, the observed SD of diastolic blood pressure (DBP) responses, and a DBP response difference of 3 mmHg, a power of 93% is obtained.
Variability of blood pressure responses
The ABP measurements were more repeatable during the placebo periods than the OBP measurements: the coefficients of variation were 3.6%/3.5% (systolic/diastolic) in ABP and 5.4%/5.2% in OBP measurements. Therefore, we have chosen ABP responses as the primary target variable in our studies. The BP responses were highly variable, confirming earlier research and clinical data. For example, the mean 24-h systolic ABP response to 50 mg of losartan was –9.1 mmHg (n = 204). The SD of responses was 6.7 mmHg, a value lower than reported for other pharmacogenetic studies with ABP measurements, and the responses ranged from + 9 (increase) to –32 (decrease) mmHg.
The within-subject correlations of BP responses between the study drugs were variable, with best correlations between bisoprolol and losartan (r = 0.32/0.39; SBP/DBP responses), and amlodipine and hydrochlorothiazide (r = 0.35/0.35), largely confirming the findings of Brown and co-workers (Citation5). The data are presented in detail in our previous paper (Citation2).
Multiple regression modelling of blood pressure responses using clinical chemical and endocrine variables
We have analysed BP responses to the four antihypertensive treatments using parameters available in clinical practice (Citation2,Citation6). Here we describe the main results of stepwise multiple regression modelling using a combined set of parameters. In this exploratory stepwise modelling, carried out with SPSS software (version 17.0, SPSS Inc., Chicago, IL, USA), a P value threshold of <0.10 was used. The 24-h ABP responses were normally distributed, or had only minor deviations not affecting the models. The baseline predictors in the analyses were transformed to normal distribution when needed and included the following: placebo 24-h ABP level (systolic or diastolic), age, earlier use of antihypertensive medication (no/yes) or a diuretic (no/yes), body mass index, waist-to-hip ratio, current smoking (no/yes), plasma renin activity (PRA), serum aldosterone, serum aldosterone-to-PRA ratio, daily urinary excretions of sodium and albumin, and serum sodium, potassium, insulin, fasting glucose, urate, cholesterol, triglycerides, calcium, and creatinine. Of highly correlated parameters, only the best predictor was included in the final model. The combined results are shown in .
Table I. Stepwise multiple regression modelling of ambulatory blood pressure responses to four antihypertensive monotherapies. P values, direction of the effect, and semipartial R2 values are shown.
Losartan
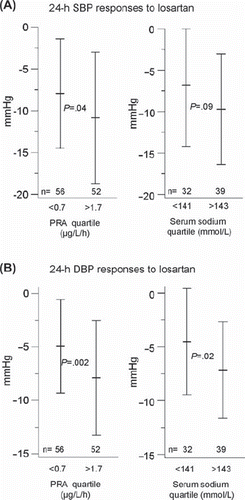
Previously proposed predictors of BP response to angiotensin-blocking agents include PRA (higher values, better response) and African-American race (lower response) (Citation7,Citation8). In the GENRES Study, significant predictors of both systolic and diastolic 24-h ABP responses to losartan were PRA, placebo BP level, and fasting serum glucose and sodium levels (). The finding of PRA as the strongest predictor (P values 0.0002/0.00001 for SBP/DBP responses in multiple regression analysis) confirms earlier reports and partially validates the design of the GENRES Study. As novel findings, lower fasting serum glucose and higher serum sodium levels predicted better response to losartan (Citation6). shows the BP responses in extreme quartiles of PRA and serum sodium levels.
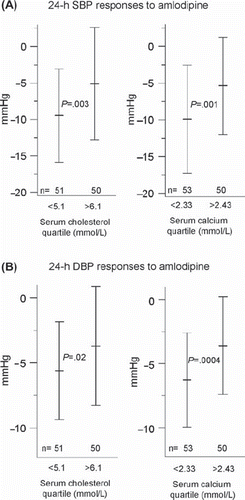
Figure 1. Ambulatory 24-h systolic (A) and diastolic (B) blood pressure responses to losartan in the lowest and the highest quartiles of plasma renin activity (PRA) and serum sodium. Means and SDs are shown. Statistical significance between the extreme quartiles was calculated with Student's t test. SBP = systolic blood pressure; DBP = diastolic blood pressure; PRA = plasma renin activity.
Bisoprolol
Higher PRA levels are known to predict better BP response to beta-blockers (Citation9), which was confirmed in our study (Citation6). In addition to placebo BP level and PRA, no other parameters predicted both systolic and diastolic ABP responses to bisoprolol. From the four study drugs, lowest R-square (the coefficient of determination) values were obtained for bisoprolol using the described model ().
Amlodipine
Higher age is known to predict better response to calcium channel blockers (Citation8), which was even observed in the GENRES Study with a relatively narrow age range (35–60 years) of subjects (). In addition, better systolic and diastolic ABP responses to amlodipine were predicted by higher placebo BP level, lower serum cholesterol, lower daily urinary excretion of sodium, and lower serum calcium (, ) (Citation2,Citation6). The associations of cholesterol and calcium have earlier been described, but only in small studies (Citation10–12).
Pharmacogenetics of blood pressure responses
Using the GENRES Study as a platform, we have analysed potential pharmacogenetic associations of several hypertension candidate genes, as also derived from a number of gene variants showing positive signals in GWAS data (Citation13–17), with BP responses to BP-lowering drugs. No significant positive results on any of the four antihypertensive drugs were obtained for polymorphisms angiotensinogen (AGT) Met235 Thr, angiotensin-converting enzyme (ACE) I/D, angiotensin type 1 receptor (AGTR1) 1166A/C, alpha-adducin (ADD1) Gly460Trp, beta-1-adrenergic receptor (ADRB1) Ser49Gly and Arg389Gly, beta-2-adrenergic receptor (ADRB2) Gly16Arg and Glu27Gln, or CYP2C9 *1/*2/*3 (Citation18–20). The negative results do not, however, exclude the possibility that these polymorphisms are related to clinical outcomes in hypertensive patients. A detailed comparison of these data to the vast field of previous pharmacogenomics studies, mostly carried out with smaller materials, single drugs, and without 24-h BP recordings, is outside the scope of this mini-review. Considering large-scale clinical studies with BP responses as one of the end-points, our results on negative findings with the ACE and ADD1 polymorphisms are in agreement with those reported from the very large GenHAT study (Citation21,Citation22). The candidate gene approach also yielded negative results in the large EUROPA/PERGENE study, where no significant predictors of BP response to an ACE inhibitor among 52 SNPs in 12 genes within the renin-angiotensin-aldosterone system were found (Citation23).
As the next step we proceeded to analysis of variations identified in the large GWAS of population BP levels (Citation13–17). We selected 19 SNPs (Citation24) and analysed their association with BP responses to all four study drugs, which resulted in 76 comparisons (counting the highly correlated SBP and DBP responses as one) and a Bonferroni-corrected P value requirement of <0.0007 for statistical significance (considering the SBP and DBP responses uncorrelated would give 152 comparisons and a P level requirement of <0.0003). In addition to some suggestive findings, one significant association was found. An intronic STK39 SNP (rs6749447) was associated with 24-h ABP response to losartan, with P values of 0.0005/0.0002 (systolic/diastolic) in multivariate analysis (Citation24). Between the extreme genotype groups, the differences of mean systolic/diastolic 24-h ABP responses were 3.3/2.6 mmHg. The association, although not replicated thus far, is physiologically plausible since the STK39-encoded kinase SPAK is involved in the activation of renal ion transporters by angiotensin II (Citation25–27). Although the thiazide-sensitive sodium-chloride cotransporter is also regulated by SPAK, the two analysed STK39 SNPs were not associated with BP response to hydrochlorothiazide (Citation24), in congruence with data from another report (Citation28).
Conclusions
BP responses to antihypertensive drugs display marked interindividual variability. Several baseline demographic and laboratory parameters predict BP responses to some extent, but classification of patients according to these variables would leave significant overlap in BP responses, well demonstrated by the SD bars of the BP responses (). Pretreatment profiling of patients may decrease the probability of treatment failure (Citation29), but also the non-preferred drugs should be considered at a later stage if target BP levels are not reached or adverse effects occur when the primary alternative is used as antihypertensive drug. At present, there is insufficient evidence to support the use of genetic polymorphisms as predictors of BP response in essential hypertension. It is still possible that large-scale genomic techniques may help to detect novel pathways of BP regulation and thereby also identify new targets for pharmacogenomics approaches.
Declaration of interest: The authors declare no conflict of interest.
References
- Attwood S, Bird R, Burch K, Casadei B, Coats A, Conway J, . Within-patient correlation between the antihypertensive effects of atenolol, lisinopril and nifedipine. J Hypertens. 1994;12:1053–60.
- Hiltunen TP, Suonsyrjä T, Hannila-Handelberg T, Paavonen KJ, Miettinen HE, Strandberg T, . Predictors of antihypertensive drug responses: initial data from a placebo-controlled, randomized, cross-over study with four antihypertensive drugs (The GENRES Study). Am J Hypertens. 2007;20:311–8.
- Lloyd-Jones DM, Evans JC, Larson MG, O'Donnell CJ, Roccella EJ, Levy D. Differential control of systolic and diastolic blood pressure: factors associated with lack of blood pressure control in the community. Hypertension. 2000;36:594–9.
- Varis J, Savola H, Vesalainen R, Kantola I. Treatment of hypertension in Finnish general practice seems unsatisfactory despite evidence-based guidelines. Blood Press. 2009;18:62–7.
- Deary AJ, Schumann AL, Murfet H, Haydock SF, Foo RS, Brown MJ. Double-blind, placebo-controlled crossover comparison of five classes of antihypertensive drugs. J Hypertens. 2002;20:771–7.
- Suonsyrjä T, Hannila-Handelberg T, Paavonen KJ, Miettinen HE, Donner K, Strandberg T, . Laboratory tests as predictors of the antihypertensive effects of amlodipine, bisoprolol, hydrochlorothiazide and losartan in men: results from the randomized, double-blind, crossover GENRES Study. J Hypertens. 2008;26:1250–6.
- Laragh JH, Letcher RL, Pickering TG. Renin profiling for diagnosis and treatment of hypertension. JAMA. 1979;241:151–6.
- Materson BJ, Reda DJ, Cushman WC, Massie BM, Freis ED, Kochar MS, . Single-drug therapy for hypertension in men. A comparison of six antihypertensive agents with placebo. The Department of Veterans Affairs Cooperative Study Group on Antihypertensive Agents. N Engl J Med. 1993;328:914–21.
- Preston RA, Materson BJ, Reda DJ, Williams DW, Hamburger RJ, Cushman WC, . Age-race subgroup compared with renin profile as predictors of blood pressure response to antihypertensive therapy. JAMA. 1998;280: 1168–72.
- Mazeaud MM, Sang KH, Astarie C, Levenson J, Simon A, Devynck MA. Hypercholesterolemia modulates the effects of nitrendipine on blood pressure and platelet function in essential hypertension. J Cardiovasc Pharmacol. 1991; 18(Suppl 10):S46–51.
- Resnick LM, Nicholson JP, Laragh JH. Calcium, the renin-aldosterone system, and the hypotensive response to nifedipine. Hypertension. 1987;10:254–8.
- Midtbø K, Hals O. Serum ionized calcium—a predictor of therapeutic response to slow calcium channel blockade in essential hypertension. Angiology. 1987;38:841–6.
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78.
- Wang Y, O'Connell JR, McArdle PF, Wade JB, Dorff SE, Shah SJ, . Whole-genome association study identifies STK39 as a hypertension susceptibility gene. Proc Natl Acad Sci U S A. 2009;106:226–31.
- Org E, Eyheramendy S, Juhanson P, Gieger C, Lichtner P, Klopp N, . Genome-wide scan identifies CDH13 as a novel susceptibility locus contributing to blood pressure determination in two European populations. Hum Mol Genet. 2009;18:2288–96.
- Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, . Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–87.
- Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, . Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–76.
- Suonsyrjä T, Hannila-Handelberg T, Fodstad H, Donner K, Kontula K, Hiltunen TP. Renin-angiotensin system and alpha-adducin gene polymorphisms and their relation to responses to antihypertensive drugs: results from the GENRES study. Am J Hypertens. 2009;22:169–75.
- Suonsyrjä T, Donner K, Hannila-Handelberg T, Fodstad H, Kontula K, Hiltunen TP. Common genetic variation of beta1- and beta2-adrenergic receptor and response to four classes of antihypertensive treatment. Pharmacogenet Genomics. 2010;20:342–5.
- Donner KM, Hiltunen TP, Suonsyrjä T, Hannila-Handelberg T, Tikkanen I, Antikainen M, . CYP2C9 genotype modifies activity of the renin-angiotensin-aldosterone system in hypertensive men. J Hypertens. 2009;27:2001–9.
- Arnett DK, Davis BR, Ford CE, Boerwinkle E, Leiendecker-Foster C, Miller MB, . Pharmacogenetic association of the angiotensin-converting enzyme insertion/deletion polymorphism on blood pressure and cardiovascular risk in relation to antihypertensive treatment: the Genetics of Hypertension-Associated Treatment (GenHAT) study. Circulation. 2005;111:3374–83.
- Davis BR, Arnett DK, Boerwinkle E, Ford CE, Leiendecker-Foster C, Miller MB, . Antihypertensive therapy, the alpha-adducin polymorphism, and cardiovascular disease in high-risk hypertensive persons: the Genetics of Hypertension-Associated Treatment Study. Pharmacogenomics J. 2007;7: 112–22.
- Brugts JJ, Isaacs A, de Maat MP, Boersma E, van Duijn CM, Akkerhuis KM, . A pharmacogenetic analysis of determinants of hypertension and blood pressure response to angiotensin-converting enzyme inhibitor therapy in patients with vascular disease and healthy individuals. J Hypertens. 2011;29:509–19.
- Donner KM, Hiltunen TP, Hannila-Handelberg T, Suonsyrjä T, Kontula K. STK39 variation predicts ambulatory blood pressure response to losartan in hypertensive men. Hypertension Res. 2012;35:107–14.
- San-Cristobal P, Pacheco-Alvarez D, Richardson C, Ring AM, Vazquez N, Rafiqi FH, . Angiotensin II signaling increases activity of the renal Na-Cl cotransporter through a WNK4-SPAK-dependent pathway. Proc Natl Acad Sci U S A. 2009;106:4384–9.
- Talati G, Ohta A, Rai T, Sohara E, Naito S, Vandewalle A, . Effect of angiotensin II on the WNK-OSR1/SPAK-NCC phosphorylation cascade in cultured mpkDCT cells and in vivo mouse kidney. Biochem Biophys Res Commun. 2010;393:844–8.
- Rafiqi FH, Zuber AM, Glover M, Richardson C, Fleming S, Jovanović S, . Role of the WNK-activated SPAK kinase in regulating blood pressure. EMBO Mol Med. 2010; 2:63–75.
- Duarte JD, Lobmeyer MT, Wang Z, Chapman AB, Gums JG, Langaee TY, . Lack of association between polymorphisms in STK39, a putative thiazide response gene, and blood pressure response to hydrochlorothiazide. Pharmacogenet Genomics. 2010;20:516–9.
- Alderman MH, Cohen HW, Sealey JE, Laragh JH. Pressor responses to antihypertensive drug types. Am J Hypertens. 2010;23:1031–7.