Abstract
Adiponectin (ADIPOQ) plays an important role in the pathogenesis of diabetic nephropathy (DN) and previous studies regarding the association between ADIPOQ polymorphisms and DN risk reported conflicting results. To derive a more precise estimation of this association, we performed a meta-analysis to assess the association between four ADIPOQ polymorphisms [−11391G > A (rs17300539), −11377C > G (rs266729), +45T > G (rs2241766), and +276G > T (rs1501299)] and risk for DN. Odds ratios (ORs) with corresponding 95% confidence intervals (95% CIs) were pooled to assess the association between four aforementioned polymorphisms and susceptibility to DN. Based on the included criteria, we selected 13 articles, among which 7 studies (cases/controls: 2749/7585) for −11391G > A, 8 studies for −11377C > G (3074/3842), 9 studies for +45T > G (2654/7710), and 10 studies for +276G > T (2812/7821), respectively. Our meta-analysis indicated no evidence heterogeneity among the included studies; thus, the fixed-effects model was used. Overall, there was an association between ADIPOQ −11391A allele with increased DN risk (OR = 1.186, 95% CI: 1.051–1.338, p = 0.006). Subgroup by ethnicity suggested significant association between +45T > G polymorphism and DN risk among Caucasians (OR = 1.122, 95% CI: 1.007–1.250, p = 0.038). Sensitivity analysis suggested exclusion of any single study did not materially alter the overall pooled ORs above. Future studies are needed to validate these findings.
Introduction
Diabetic nephropathy (DN) is a common microvascular complication of type 1 and type 2 diabetes (T1DM and T2DM), and it is the primary cause of end-stage renal disease (ESRD) worldwide.Citation1–3 DN results from various causes including genetic and environmental factors, and major genes that contribute to the etiology of DN have yet to be identified.Citation4–8 The adiponectin gene (gene ID 9370) is encoded as ADIPOQ (adipocyte C1q and collagen domain containing); and GBP28, ACRP30, APM1, and ACDC are this gene’s alternative names.Citation9 The ADIPOQ gene is located in chromosome 3q27 and consists of three exons and two introns.Citation9–11 ADIPOQ is an adipokine, which not only performs an important role in the regulation of insulin action, glucose, and lipid metabolism but also exerts anti-inflammatory and anti-atherogenic effects, with a low circulating concentration in cases of insulin resistance, T2DM, and coronary heart disease.Citation12–15 In contrast, ADIPOQ levels are high in cases of DN, but it is still unclear whether these high levels are a cause or a consequence of the disease.Citation16 The influence of ADIPOQ genetic polymorphisms in the development of DN is, nonetheless, not fully understood. Therefore, it is urgent to elucidate the association of ADIPOQ genetic polymorphisms and susceptibility to DN. Several polymorphisms in ADIPOQ gene have been identified, with −11391G > A (rs17300539), −11377C > G (rs266729), +45T > G (rs2241766), and +276G > T (rs1501299) and DN risk being extensively evaluated.Citation8–10,Citation16–33 Evidences have suggested that genetic variations of −11391G > A and −11377C > G in the promoter region of this gene can play a vital role in the increased risk of developing nephropathy partly through affecting the plasma levels of ADIPOQ.Citation8,Citation16 In addition, genetic association studies have demonstrated that single nucleotide polymorphisms (SNPs) +45T > G in exon2 and +276G > T in intron 2 of this gene confer the risk susceptibility to the development of DN.Citation8,Citation16,Citation21 However, results are inconsistent and firm association has not yet been established.
Considering the important roles of the ADIPOQ gene to the risk of developing DN and the insufficient power of a single study to provide responsible conclusion, we conducted a meta-analysis of all available studies relating four aforementioned polymorphisms in ADIPOQ gene to shed some light on these controversial issues.
Methods
Search strategy
A comprehensive search strategy was conducted towards the electronic databases including PubMed, Embase, Medline and CNKI databases. The following search terms were used: (1) adiponectin, ADIPOQ, APM1, and ACDC; (2) diabetic nephropathy and diabetes nephropathy; and (3) polymorphism, variation, variant, and mutation. An upper date limit of August 2013 was applied and we used no lower date limit. The language was restricted to English and Chinese. In addition, we reviewed the reference lists of retrieved papers and recent reviews.
Study selection
We first performed an initial screening of titles or abstracts. A second screening was based on full-text review. The inclusion criteria were: (1) case--control or cohort studies which evaluated the association between ADIPOQ −11391G > A (rs17300539), −11377C > G (rs266729), +45T > G (rs2241766), and +276G > T (rs1501299) polymorphisms and DN risk in type 1 or type 2 diabetic patients; (2) the controls were diabetic individuals without DN; (3) used an unrelated case–control or cohort design and had available genotype/allele counts for estimating an OR with its 95% CI. If two or more studies described outcomes among the same or overlapping groups of cases or controls, only the one with the largest available data was included in the meta analysis.
Data extraction
All data were reviewed and extracted independently by two investigators. From each study the following information was extracted: ADIPOQ polymorphisms, first author, publication year, ethnicity of study populations, study design, types of diabetes, genotyping methods, the number of cases and controls, minor allele frequency (MAF) in cases and controls. Any discrepancies in the extracted data were settled by discussion and, when necessary, adjudicated by a third reviewer.
Statistical analysis
In view of articles providing data only on allele counts accounted for the majority of the included ones, and to enhance study power to detect an association, we exclusively took account of allelic model in this meta-analysis. To test the population stratification in the controls, the departure of frequencies of ADIPOQ polymorphisms using the chi-square test from expectation under Hardy–Weinberg equilibrium (HWE) was assessed in controls. Pooled ORs (95% CI) for DN risk associated with ADIPOQ gene −11391A, −11377G, +45G, and +276T alleles compared with the alternative alleles were calculated, respectively. In this analysis, both the chi-square based Q statistic test (Cochran's Q statistic) and the I2 statistic were applied to assess the between-study heterogeneity more precisely.Citation35,Citation36 Heterogeneity was considered significant for P Cochran's Q statistic <0.10. With evidence heterogeneity among studies, the random-effects model (the DerSimonian and Laird method) was used to assess the summary ORs of each study;Citation37 Otherwise, the fixed-effects model (the Mantel–Haenszel method) was used to pool the results.Citation38 For exploring between-study heterogeneity or indicating that our results were statistically robust, subgroup analysis was also performed by ethnicity for ADIPOQ −11391G > A, −11377C > G, +45T > G, and +276G > T polymorphisms. Furthermore, we conducted a sensitivity analysis to examine the influence of a single study on the overall risk estimate by omitting one study in each turn.Citation39 Eventually, potential publication bias was tested by Begg’s funnel plot and Egger’s test. Possible publication bias was assessed by visual inspection of the funnel plots in which the log ORs were plotted against their SEs.Citation40 Significance was judged at the p < 0.1 level of Egger’s test.Citation41 All analyses were performed using STATA version 12.0 (StataCorp LP, College Station, TX). p < 0.05 should be considered statistical significance, except where otherwise specified.
Results
Characteristics of included studies
A flow diagram schematizing the study selection process was shown in . Following our search strategy 397 individual abstracts were retrieved originally, and 23 full-text articles were preliminarily identified for further detailed evaluation.Citation8–10,Citation16–34,Citation42 On the basis of the exclusion criteria, 10 articles were excluded including one study containing overlapping data,Citation31 one review article,Citation34 one article with non-diabetic controls,Citation27 one article with cross-sectional in design,Citation28 one article with no included polymorphisms,Citation32 and five articles with missing relevant data.Citation10,Citation29,Citation30,Citation33,Citation42 Since the article by Vionnet et al.Citation8 categorized data in Denmark, Finland, and France populations, we extracted them as three individual case–control studies. Finally, we included 13 articles, among which 7 studies (cases/controls: 2749/7585) for −11391G > A, 8 studies for −11377C > G (3074/3842), 9 studies for +45T > G (2654/7710), and 10 studies for +276G > T (2812/7821), respectively.
The characteristics of included studies are summarized in . In the included studies, the distribution of genotypes in the controls was all in agreement with HWE, except for the +45T > G polymorphism in one studyCitation17 and the +276G > T polymorphism in one study.Citation26
Table 1. Characteristics of studies included in the meta-analysis.
Meta-analysis results
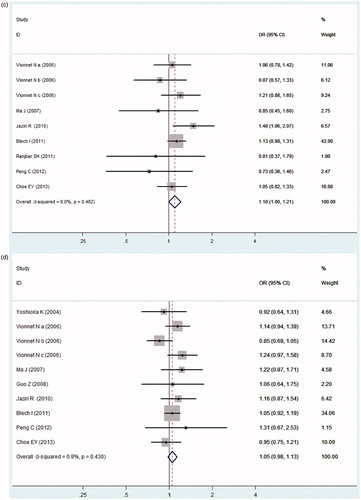
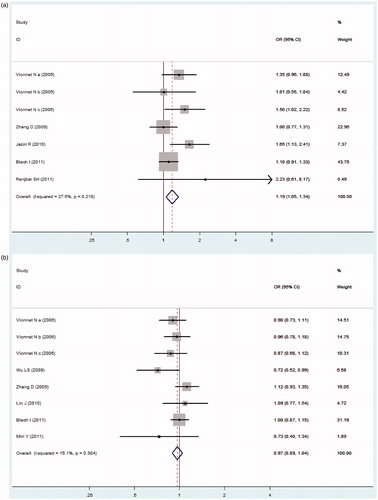
In , our meta-analysis indicated no obvious heterogeneity among the included studies of the ADIPOQ −11391G > A polymorphism (Pheterogeneity = 0.218, I2 = 27.6%), −11377C > G polymorphism (Pheterogeneity = 0.304, I2 = 16.1%), +45T > G polymorphism (Pheterogeneity = 0.482, I2 = 0.0%), and +276G > T polymorphism (Pheterogeneity = 0.430, I2 = 0.9%), thus the fixed-effects model was used. ADIPOQ −11391A allele was associated with increased DN risk (OR = 1.186, 95% CI: 1.051–1.338, p = 0.006). However, no significant association between the −11377C > G, +45T > G or +276G > T polymorphism and DN risk was found in our present study (OR = 0.966, 95% CI: 0.894–1.044, p = 0.387; OR = 1.100, 95% CI: 0.997–1.213, p = 0.057; OR = 1.051, 95% CI: 0.976–1.133, p = 0.189; respectively, ).
Figure 2. Forest plot for meta-analysis association between the ADIPOQ polymorphisms with DN risk (allele model). (a) −11391G > A, (b) −11377C > G, (c) +45T > G, (d) +276G > T.
MAF of the three ADIPOQ polymorphisms among the controls across the different ethnicities (Asians and Caucasians) was also assessed (), except for −11391G > A polymorphism because all ethnicities of the included studies of this polymorphism were Caucasians. For the −11377C > G and +276G > T polymorphisms, there were similar results in the MAF among the controls across Asians and Caucasians. However, we found that the +45G allele frequency in Asian populations was 29.5% (95% CI = 23.1–35.9%), which was significantly higher than that in Caucasian populations (11.9%, 95% CI = 7.0–16.7%, p = 0.003; ).
Table 2. MAF among controls stratified by ethnicity.
Subgroup analyses
The subgroup analysis was undertaken according to ethnicity. we found that +45T > G polymorphism was significantly associated with DN in Caucasian populations (G versus T: Pheterogeneity = 0.442, I2 = 0.0%, OR = 1.122, 95% CI: 1.007–1.250, p = 0.038); for Asian populations, we did not find the similar result. However, no significant association between the −11377C > G or +276G > T polymorphism and risk of DN was found among Caucasian populations and Asian populations ().
Table 3. Stratified analyses of the ADIPOQ polymorphisms on DN risk.
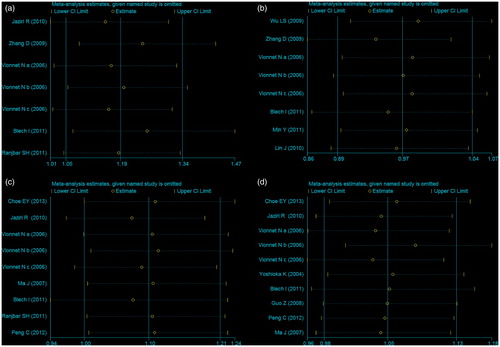
Sensitivity analysis
In this meta-analysis, we also performed sensitivity analyses to test the stability of the overall results. By exclusion of the study violating HWE or altering the statistic models, no material alternation was detected ().
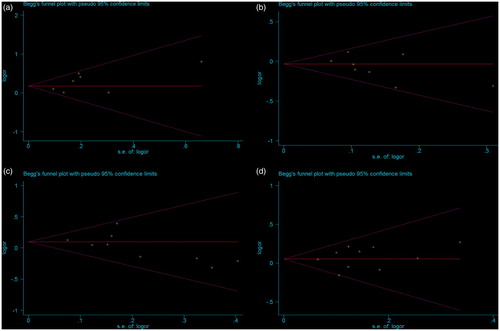
Publication bias
Funnel plot and Egger's test were conducted to evaluate the publication bias in this meta-analysis. In , the shape of the funnel plot did not reveal obvious asymmetry. Then, Egger’s linear regression test was used to provide statistical evidence of funnel plots asymmetry. For the ADIPOQ polymorphisms −11391G > A, −11377C > G, +45T > G, and +276G > T, the p values of Egger’s linear regression test were 0.197, 0.182, 0.171, and 0.599, respectively. Hence, the above results indicated that publication bias was not evident in this meta-analysis.
Discussion
Diabetes mellitus affects the kidney in stages, and 30–40% of the patients with T1DM and T2DM suffer from DN.Citation1,Citation2,Citation43 It is well recognized that DN has genetic components and individual susceptibility, while the pathophysiology of DN remains unclear and needs to be fully investigated.Citation6,Citation44–47 Recently, more attention has been paid to the identification of the genetic variations of nephropathy in T1DM and T2DM. Thus, identification of susceptibility or resistance genes in DN will provide better knowledge of pathomechanism, genetic biomarkers and future therapies.Citation48–50
Considerable studies have indicated that the levels of ADIPOQ are influenced by genetic polymorphisms in the ADIPOQ geneCitation51–54 and these polymorphisms have been implicated in DN.Citation8–10,Citation16–32 Nephropathy has been associated with high ADIPOQ levels in T1DM and T2DM.Citation16,Citation31 Thus, the genetic studies of DN and ADIPOQ gene will give new insight into the underlying etiology and lead to new therapies for treatment and prevention in this disease. Previous studies regarding the association between SNPs within ADIPOQ and DN risk reported conflicting results. Studies proposed that these SNPs were determinants of the DN risk.Citation9,Citation16,Citation21 However, several studies also showed that these SNPs were not associated with susceptibility to DN.Citation17,Citation22,Citation23 Among these SNPs, −11391G/A and −11377C/G are located in the ADIPOQ gene promoter region, whereas +45T > G and +276G > T are located in the exon2 and intron 2 region.Citation9–11 Evidences suggested that genetic variation in the promoter region of this gene could play an important role in the risk of DN partly through affecting the plasma levels of ADIPOQ.Citation8,Citation16 Moreover, studies demonstrated that −11377C > G polymorphism could alter the sequence in one of four SP1 binding sites in this promoter region and reduce ADIPOQ promoter transcription activity.Citation9,Citation55 Jaziri et al.Citation16 reported high ADIPOQ concentrations associated with the −11391A, +45G alleles might be the cause, rather than a consequence of DN. Several studies indicated that SNP +276G > T was associated with lower plasma ADIPOQ levels, while no significant association between +276G > T and the prevalence of DN was found in other studies.Citation17,Citation20,Citation23 In view of inconsistent results, we performed a meta-analysis of these SNPs to derive a more precise estimation of the relationship between these SNPs and susceptibility to DN.
In this meta-analysis, 14 articles (7 studies for −11391G > A, 8 studies for −11377C > G, 9 studies for +45T > G, and 10 studies for +276G > T) were performed to provide the assessment of the relationship between ADIPOQ polymorphisms and DN. No obvious heterogeneity was found among the included studies. For ADIPOQ −11391G > A polymorphism, studies observed significant association with DN risk in the allele genetic comparison model. For the −11377C > G, +45T > G, or +276G > T polymorphism, no significant association with DN was found. However, in the subgroup analysis of +45T > G by ethnicity, the results demonstrated significant association was found among Caucasian populations, partly because there was evidence difference in the MAF of +45T > G among the controls across Asians and Caucasians. Sensitivity analysis indicated that our results were statistically robust.
Some possible limitations of this meta-analysis should be acknowledged in explaining the results. First, most included studies were retrospective in design. Second, publication bias might have occurred because only published studies were included and unpublished studies which had null results were missed, which might bias the results. Therefore, these biases could lead to an overestimation of the effects in this meta-analysis. Third, the studies involved were relatively less and small study samples. More and larger studies should be warranted in future to elucidate the role of the variant in DN susceptibility. Finally, DN is a multifactorial disease that results from complex interactions between many environmental and genetic factors. We only focused on ADIPOQ gene polymorphisms, and did not cover other DN susceptibility genes or polymorphisms.
In conclusion, for the −11391G > A and +45T > G polymorphisms, significant association with DN was found by the allele genetic comparison model in overall or subgroup analysis. However, our meta-analysis provided evidence that −11377C > G and +276G > T polymorphisms were unlikely to be associated with genetic susceptibility of DN based on the current published studies. To get a more exact conclusion on the effects of ADIPOQ polymorphisms on DN risk, future large and well-designed studies are necessary.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Remuzzi G, Schieppati A, Ruggenenti P. Clinical practice. Nephropathy in patients with type 2 diabetes. N Engl J Med. 2002;346(15):1145–1151
- Tahrani AA, Bailey CJ, Del Prato S, Barnett AH. Management of type 2 diabetes: new and future developments in treatment. Lancet. 2011;378(9786):182–197
- Nyenwe EA, Jerkins TW, Umpierrez GE, Kitabchi AE. Management of type 2 diabetes: evolving strategies for the treatment of patients with type 2 diabetes. Metabolism. 2011;60(1):1–23
- Maeda S, Imamura M, Kurashige M, et al. Replication study for the association of 3 SNP loci identified in a genome-wide association study for diabetic nephropathy in European type 1 diabetes with diabetic nephropathy in Japanese patients with type 2 diabetes. Clin Exp Nephrol. 2013 . [Epub ahead of print]. doi: 10.1007/s10157-013-0797-5
- Yang S, Zhang J, Feng C, Huang G. MTHFR 677T variant contributes to diabetic nephropathy risk in Caucasian individuals with type 2 diabetes: a meta-analysis. Metabolism. 2013;62(4):586–594
- Sandholm N, Salem RM, McKnight AJ, et al. New susceptibility loci associated with kidney disease in type 1 diabetes. PLoS Genet. 2012;8(9):e1002921
- Karadeniz M, Erdogan M, Cetinkalp S, Berdeli A, Eroglu Z, Ozgen AG. Monocyte chemoattractant protein-1 (MCP-1) 2518G/A gene polymorphism in Turkish type 2 diabetes patients with nephropathy. Endocrine. 2010;37(3):513–517
- Vionnet N, Tregouët D, Kazeem G, et al. Analysis of 14 candidate genes for diabetic nephropathy on chromosome 3q in European populations: strongest evidence for association with a variant in the promoter region of the adiponectin gene. Diabetes. 2006;55(11):3166–3174
- Zhang D, Ma J, Brismar K, Efendic S, Gu HF. A single nucleotide polymorphism alters the sequence of SP1 binding site in the adiponectin promoter region and is associated with diabetic nephropathy among type 1 diabetic patients in the Genetics of Kidneys in Diabetes Study. J Diabetes Complications. 2009;23(4):265–272
- Zhang D, Efendic S, Brismar K, Gu HF. Effects of MCF2L2, ADIPOQ and SOX2 genetic polymorphisms on the development of nephropathy in type 1 Diabetes Mellitus. BMC Med Genet. 2010;11:116
- Chu H, Wang M, Zhong D, et al. AdipoQ polymorphisms are associated with type 2 diabetes mellitus: a meta-analysis study. Diabetes Metab Res Rev. 2013;29(7):532–545
- Qi L, Li T, Rimm E, et al. The +276 polymorphism of the APM1 gene, plasma adiponectin concentration, and cardiovascular risk in diabetic men. Diabetes. 2005;54(5):1607–1610
- Francke S, Manraj M, Lacquemant C, et al. A genome-wide scan for coronary heart disease suggests in Indo-Mauritians a susceptibility locus on chromosome 16p13 and replicates linkage with the metabolic syndrome on 3q27. Hum Mol Genet. 2001;10(24):2751–2765
- Fumeron F, Aubert R, Siddiq A, et al. Epidemiologic Data on the Insulin Resistance Syndrome (DESIR) Study Group: adiponectin gene polymorphisms and adiponectin levels are independently associated with the development of hyperglycemia during a 3-year period: the epidemiologic data on the insulin resistance syndrome prospective study. Diabetes. 2004;53(4):1150–1157
- Menzaghi C, Trischitta V, Doria A. Genetic influences of adiponectin on insulin resistance, type 2 diabetes, and cardiovascular disease. Diabetes. 2007;56(5):1198–1209
- Jaziri R, Aubert R, Roussel R, et al. Association of ADIPOQ genetic variants and plasma adiponectin isoforms with the risk of incident renal events in type 2 diabetes. Nephrol Dial Transplant. 2010;25(7):2231–2237
- Choe EY, Wang HJ, Kwon O, et al. Variants of the adiponectin gene and diabetic microvascular complications in patients with type 2 diabetes. Metabolism. 2013;62(5):677–685
- Wu LS, Hsieh CH, Pei D, Hung YJ, Kuo SW, Lin E. Association and interaction analyses of genetic variants in ADIPOQ, ENPP1, GHSR, PPARgamma and TCF7L2 genes for diabetic nephropathy in a Taiwanese population with type 2 diabetes. Nephrol Dial Transplant. 2009;24(11):3360–3366
- Ma J, Möllsten A, Falhammar H, et al. Genetic association analysis of the adiponectin polymorphisms in type 1 diabetes with and without diabetic nephropathy. J Diabetes Complications. 2007;21(1):28–33
- Yoshioka K, Yoshida T, Umekawa T, et al. Adiponectin gene polymorphism (G276T) is not associated with incipient diabetic nephropathy in Japanese type 2 diabetic patients. Metabolism. 2004;53(9):1223–1226
- Blech I, Katzenellenbogen M, Katzenellenbogen A, et al. Predicting diabetic nephropathy using a multifactorial genetic model. PLoS One. 2011;6(4):e18743
- Ranjbar SH, Amoli MM, Sajadi M, et al. Genetic association analysis of the adiponectin polymorphisms in type 2 diabetes with and without complications. Iran J Diabetes Lipid Disord. 2011;10:1–4
- Guo Z, Wu S, Sun Y. Relationship between single nucleotide polymorphism at position 276 in adiponectin gene and type 2 diabetic nephropathy. J Shanxi Med Univ. 2008;39(6):507–510. [in Chinese]
- Min Y, Chen J, Zhu L, Li X, Wei J, Su K. Association of SNP-11377C/G in proximal promoter region of adiponectin gene with type 2 diabetic nephropathy. Shandong Med J. 2011;5:65–66. [in Chinese]
- Lin J, Shi Y, Li Y, Tao G. Association of gene polymorphism in promoter region of adiponectin gene and albuminuria in patients with type 2 diabetes mellitus. Chin J Diabetes Mellitus. 2010;2(2):106–110. [in Chinese]
- Peng C, Hong Y, Fu L. Association of adiponectin gene polymorphism in type 2 diabetes with nephropathy. J Med Res. 2012;41(4):156–159. [in Chinese]
- Bostrom MA, Freedman BI, Langefeld CD, Liu L, Hicks PJ, Bowden DW. Association of adiponectin gene polymorphisms with type 2 diabetes in an African American population enriched for nephropathy. Diabetes. 2009;58(2):499–504
- Prior SL, Javid J, Gill GV, Bain SC, Stephens JW. The adiponectin rs17300539 G > A variant and nephropathy risk. Kidney Int. 2008;74(10):1361
- Zhao H, Chen L, Zhang C, Han J. Polymorphism in the 5′promoter region of the APN gene contributes to development of type 2 diabetes nephropathy. Proc Clin Med. 2008;1(7):483–485. [in Chinese]
- Jia M, Feng X, Zhang Z, Qiao B, Qin X, Sun Y. Correlation of polymorphism of adiponectin promoter with diabetes type 2 and its complications. Chin J Lab Med. 2008;31(2):163–169. [in Chinese]
- Jorsal A, Tarnow L, Frystyk J, et al. Serum adiponectin predicts all-cause mortality and end stage renal disease in patients with type I diabetes and diabetic nephropathy. Kidney Int. 2008;74(5):649–654
- Rudofsky G Jr, Schlimme M, Schlotterer A, et al. No association of the 94T/G polymorphism in the adiponectin gene with diabetic complications. Diabetes Obes Metab. 2005;7(4):455–459
- Gu HF, Alvarsson A, Efendic S, Brismar K. SOX2 has gender-specific genetic effects on diabetic nephropathy in samples from patients with type 1 diabetes mellitus in the GoKinD study. Gend Med. 2009;6(4):555–564
- Gu HF. Biomarkers of adiponectin: plasma protein variation and genomic DNA polymorphisms. Biomark Insights. 2009;4:123–133
- Higgins JPT, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560
- Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10:101–129
- DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188
- Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748
- Tobias A. Assessing the influence of a single study in the meta-analysis estimate. Stata Tech Bull. 1999;8:15–17
- Stuck AE, Rubenstein LZ, Wieland D. Bias in meta-analysis detected by a simple, graphical test. Asymmetry detected in funnel plot was probably due to true heterogeneity. BMJ. 1998;316(7129):469--471
- Bowden J, Tierney JF, Copas AJ, Burdett S. Quantifying, displaying and accounting for heterogeneity in the meta-analysis of RCTs using standard and generalised Q statistics. BMC Med Res Methodol. 2011;11:41
- Chung HF, Chen PS, Long K, Hsu CC, Huang MC. The association of ADIPOQ gene polymorphisms and clinical risk factors with nephropathy progression in type 2 diabetes. Meeting Abstracts. Experimental Biology 2012, EB, San Diego, CA, Apr 21–25, 2012
- Hasslacher C, Ritz E, Wahl P, Michael C. Similar risks of nephropathy in patients with type I or type II diabetes mellitus. Nephrol Dial Transplant. 1989;4:859–863
- Makita Y, Moczulski DK, Bochenski J, Smiles AM, Warram JH, Krolewski AS. Methylenetetrahydrofolate reductase gene polymorphism and susceptibility to diabetic nephropathy in type 1 diabetes. Am J Kidney Dis. 2003;41(6):1189–1194
- Wang L, Teng Z, Cai S, Wang D, Zhao X, Yu K. The association between the PPARγ2 Pro12Ala polymorphism and nephropathy susceptibility in type 2 diabetes: a meta-analysis based on 9,176 subjects. Diagn Pathol. 2013;8(1):118 (1--7). doi: 10.1186/1746-1596-8-118
- Freedman BI, Tuttle AB, Spray BJ. Familial predisposition to nephropathy in African-Americans with non-insulin-dependent diabetes mellitus. Am J Kidney Dis. 1995;25(5):710–713
- Österholm AM, He B, Pitkaniemi J, et al. Genome-wide scan for type 1 diabetic nephropathy in the Finnish population reveals suggestive linkage to a single locus on chromosome 3q. Kidney Int. 2007;71(2):140–145
- Freedman BI, Bostrom M, Daeihagh P, Bowden DW. Genetic factors in diabetic nephropathy. Clin J Am Soc Nephrol. 2007;2(6):1306–1316
- McKillop AM, Flatt PR. Recent developments in the search for biomarkers for the diagnosis and monitoring of diabetes. Recent Pat Endocr Metab Immune Drug Discov. 2008;2(3):172–177
- Yamagishi SI, Fukami K, Ueda S, Okuda S. Molecular mechanisms of diabetic nephropathy and its therapeutic intervention. Curr Drug Targets. 2007;8(8):952–959
- Jee SH, Sull JW, Lee JE, et al. Adiponectin concentrations: a genome-wide association study. Am J Hum Genet. 2010;87(4):545–552
- Ling H, Waterworth DM, Stirnadel HA, et al. Genome-wide linkage and association analyses to identify genes influencing adiponectin levels: the GEMS Study. Obesity. 2009;17(4):737–744
- Richards JB, Waterworth D, O'Rahilly S, et al. A genome-wide association study reveals variants in ARL15 that influence adiponectin levels. PLoS Genet. 2009;5(12):e1000768
- Heid IM, Wagner SA, Gohlke H, et al. Genetic architecture of the APM1 gene and its influence on adiponectin plasma levels and parameters of the metabolic syndrome in 1,727 healthy Caucasians. Diabetes. 2006;55(2):375–384
- Laumen H, Saningong AD, Heid IM, et al. Functional characterization of promoter variants of the adiponectin gene complemented by epidemiological data. Diabetes. 2009;58(4):984–991