Abstract
To clarify the effect of nitric oxide synthase (NOS) type III 4b/a polymorphism on the susceptibility to diabetic nephropathy (DN) by meta-analysis, we performed a computerized search of PubMed, EMBASE, Chinese BioMedical Literature Database, China Science and Technology Journal Database, Chinese Journal Full-text Database and WanFang to identity case–control studies on relationship between NOS type III 4b/a polymorphism and the susceptibility to DN. Statistic analysis and heterogeneity test were conducted by StataSE12. The meta-analysis involved 26 studies for DN comparing with diabetes mellitus (DM) and 15 studies for DN comparing with healthy persons, which provided 6144/4900 cases/controls and 2134/2348 cases/controls, respectively. Moderate heterogeneity was found among including studies. The qualities of half studies are low. Meta-analysis derived a significant association between the NOS type III 4b/a and the risk of developing DN in Asian population. The sensitivity analysis (exclusion of studies not in Hardy–Weinberg equilibrium) produced non-significant changes. Compared with diabetes patients, the pre-allele model produced certain association in global populations [odds ratio (OR) = 1.26, 95% confidence interval (95% CI): 1.10–1.45], significant association in Asian population (OR = 1.51, 95% CI: 1.13–2.01) and certain association in type 2 DM patients (OR = 1.29, 95% CI: 1.09–1.54). Only in the dominant model, the funnel plot and Egger’s test provided evidence of publication bias (p = 0.024). Overall, although there is some evidence of association between NOS type III 4b/a polymorphism and DN in Asian population, the more reliable findings need further and more rigorous, prospective and high-quality studies.
Introduction
Diabetic nephropathy (DN) is one of the most devastating microvascular complications in patients with diabetes mellitus (DM). It occurs in 30%–40% of patients with type 1 diabetes mellitus (T1DM), 20–25 years after disease onset and in an increasing percentage (up to 25%) of type 2 diabetes mellitus (T2DM) patients after a variable number of years.Citation1 The typical clinical course of DN includes five consecutive stages, overt DN is strictly defined based on the existence of proteinuria (UAER >20 µg/min or AER >30 mg/d) and/or renal failure.Citation2 The risk of DN is greater when exposes to hyperglycemic environment, hyperlipidemia, hypertension and obesity. But not every DM patient develops into DN when exposes above, the specific etiology is undiscovered. Most studies indicate the development of DN is multifactorial, which involves both environmental and genetic factors, and it is widely accepted that individuals with DM may be at different levels of susceptibility to nephropathy. Several genes have been implicated in DN.Citation3,Citation4 Nevertheless, which gene is the modest risk for susceptibility of DN has not yet been interpreted. Vascular endothelial dysfunction resulting from impaired nitric oxide synthase (NOS) in the endothelial cells of blood vessels has been shown to be a crucial pathophysiologic denominator for DN.Citation5,Citation6 Three distinct isoforms of NOS have been identified in humans: neuronal NOS (nNOS or NOS1), inducible NOS (iNOS or NOS2) and endothelial NOS (eNOS or NOS3).Citation7 NOS3 are a family of enzymes catalyzing the production of nitric oxide (NO) from L-arginine. NO is an important cellular signaling molecule, it diffuses from the endothelium to the vascular smooth muscle cells, where it increases the concentration of cGMP by stimulating soluble guanylate cyclase, leading to vascular relaxation.Citation8 NOS3 is found in the glomerular afferent and efferent arterioles,Citation9 generates NO in blood vessels and is involved with regulating vascular function.Citation10 Human NOS3 is encoded by a gene located on chromosome 7q35–36 comprising a 26 exons-25 introns, and its predominant form has 133 KDa.Citation11
There are numerous studies focusing on genetic polymorphisms in NOS3 and DN. Three single-nucleotide polymorphism in the promoter region [the intron 4 27-bp repeat (4b/a), the G894T missense mutation in exon 7 and the T786C] in NOS3 have attracted much attention.Citation12–14 4b/a was reported to be associated with DN according to functional experiments, but the results of association is still controversial or inconclusive.Citation15–18 These studies were based on a limited sample size or samples of different ethnicities, genotyping procedures and so on, the controls that studies chosen were different. Therefore, the conclusion of these studies may be inadequate. To shed some light on these controversial results and better address the association between NOS3 polymorphism and DN risk, we performed a meta-analysis based on all eligible available population-based association studies relating variants of the NOS3 gene to the risk of DN. We will discuss the association when comparing disparate controls DM patients and non-DM populations (that is healthy populations). Furthermore, we will explore the association based on different ethnicity and type of DM.
Materials and methods
Search strategy
We carried out a computerized literature search of PubMed, EMBASE, Chinese BioMedical Literature Database, China Science and Technology Journal Database, Chinese Journal Full-text Database and WanFang Data by using the Boolean combinations of keywords (diabetic nephropathy* or diabetic kidney disease* or diabetic renal disease* or DN) and (Polymorphism* or SNP*) and (NOS or eNOS or NOS3 or ecNOS or NOS or 4b/a). We also searched Google complementally (http://scholar.google.com./). All references cited by identified eligible studies and previous reviews were scrutinized to find additional literatures not indexed by the database. The search strategy covered all language publications from inception to 14 October 2013.
Inclusion/exclusion criteria
Studies included had to meet all the following criteria: (1) studies examined the hypothesis that NOS3 4b/a polymorphism was associated with DN; (2) diabetic patients without nephropathy or non-DM crowd or both were chose as controls; (3) they were case–control studies determined an estimate of odds ratio (OR) together with the corresponding 95% confidence interval (95% CI); (4) studies provided genotypes or alleles distribution in both case and control groups; (5) DN diagnosed by criterion of WHO,Citation19 or met UAER >30 mg/d or AER >20 mg/min (that is stage three and above). When genotypes or alleles distribution were not available, authors were contacted to request the relevant information.
Studies excluded when they met any criteria as follows: (1) studies were repetitive or the subject groups investigated overlapped with each other; (2) studies did not focus on etiology; (3) genotypes or alleles distribution in DN were miscellaneous data contained first or second stage; and (4) the effective information were not available although contacted authors.
We conducted twice preliminary test to ensure consistency. The screenings of the abstracts/titles and full-text articles were performed by two authors independently to reduce reviewer bias and errors. Any disagreements with them were resolved by discussion or the involvement of another author.
Data extraction
We extracted the following information from each study: first author, publication year, ethnicity of subject, clinical characteristics including gender, age, body mass index (BMI), diabetes duration in case groups, systolic blood pressure (SBP) and diastolic blood pressure (DBP), type of DM, genotyping method, the source of DM/DN patient and status of non-DM controls, diagnostic criteria of DM and DN and the numbers of cases, controls and their subgroups. The percentages of genotype and allele distribution were extracted or calculated for both cases and controls, also their subgroups.
We made data extraction table and conducted twice preliminary test to ensure data’s integrity and screening’s consistency. Eligibility judgment and data extraction were recorded and carried out independently by two authors in a standardized manner. Any disagreements with them were resolved by discussion or the involvement of another author.
Quality assessment
Although there is no widely agreed quality criteria for assessing case–control studies, the Newcastle-Ottawa Scale (NOS), which had been used in training of the Cochrane Non-Randomized Studies Methods Group,Citation20 was judged by Deeks et al.Citation21 to be suitable for use in a systematic review. NOS was developed to assess the quality of case–control studies and cohort studies in the interpretation of meta-analytic results. Scoring of NOS was a “star system” and based on three broad perspectives: the selection of the study groups; the comparability of the groups; and the ascertainment of either the exposure or outcome of interest for case–control or cohort studies respectively.Citation22 There can be nine stars at most. As showed in published study, the study quality of NOS can be defined three categories: the study was considered to have high quality (low risk of bias) if it obtained seven stars or above, studies that got one or zero for selection or zero for comparability or for exposure were categorized as having low quality (high risk of bias), studies that gained in between were considered as having medium quality (moderate risk of bias).Citation23
We conducted twice preliminary test to ensure data’s integrity and screening’s consistency. Quality assessments were recorded and carried out independently by two authors. Any disagreements with them were resolved by discussion or the involvement of another author.
Data synthesis and analysis
OR and 95% CI were used to assess the strength of association between NOS3 4b/a polymorphisms and DN compared with DM and non-DM crowed for per-allele genetic model (4a vs. 4b), dominant genetic model (4aa + 4ab vs. 4bb), recessive genetic model (4aa vs. 4ab + 4bb), additive genetic model (4aa vs. 4bb), co-dominant genetic model (4ab vs. 4aa + 4bb)Citation24. Between-study heterogeneity was assessed by the inconsistency index I2 statistic (ranging from 0 to 100%). An I2 < 50% indicates that heterogeneity is minor, and we can select fixed effects model (FEM) using the Mantel-Haenszel method, otherwise random effects model (REM) using the DerSimonian and Laird method was employed.Citation25,Citation26 Satisfaction of NOS3 4b/a genotypes with Hardy-Weinberg proportions was tested by the χCitation2 test in DN group and control (DM and non-DM) groups, a p > 0.05 indicates that controls are in Hardy–Weinberg equilibrium (HWE).
When the hypothesis of homogeneity was rejected by the I2 statistic, subgroup analysis was conducted in order to explore potential moderating factors for heterogeneity.Citation25 In our study, subgroup analyses were conducted for ethnicity, type of DM, diabetes duration, report of age and BMI, genotyping method, control (DM or non-DM), source of DM/DN and status of HWE in control groups. The ethnic category based on the “nine geographic race”Citation27 and skin color, respectively.
The potential publication bias was examined by funnel plot symmetry. A symmetric funnel shape indicates that publication bias is unlikely, but an asymmetric funnel suggests the possibility of publication bias. Then Egger’s test was further used to test for publication bias objectively. The significance of the intercept was determined by the t test, and a p < 0.1 was considered significantly.Citation28 We performed sensitivity analysis to examine the effect of excluding specific studies and test the robustness of result. The analyses used the statistical software StataSE12.0 (Stata Corp LP, College Station, TX).
Results
Study selection
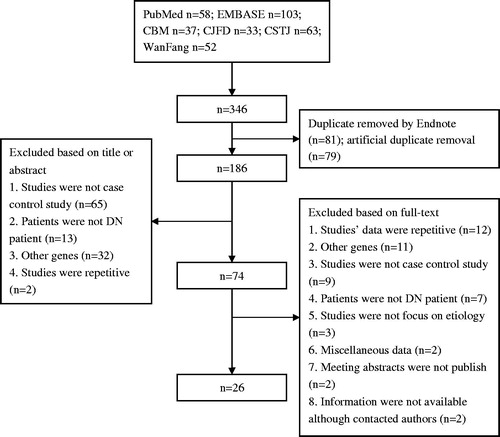
A flow chart describing the screening process is presented. As shown in , 346 references were found with our search criterion. Screened by titles and abstracts, we rejected 272 references. Based on the full-text of other 74 references, only 26 references that investigated the association between 4a/b polymorphisms and DN met our criteria.Citation15–18,Citation29–50 The most important reasons for exclusion were as follows: the subject groups investigated overlapped with each other (n = 12); the genes that focused by studies were not 4b/a (n = 11); the type of studies were not case–control studies (n = 9); and the patients that focused by studies were not patients with DN (n = 7). Other reasons included the following: studies did not focus on etiology (n = 3); genotypes or alleles distribution in DN were miscellaneous data contained first or second stage (n = 2); the effective information were not available because meeting abstracts were not published (n = 2); and authors were not contacted (n = 2). We did not find any references that met our inclusion and exclusion criteria through Google and scrutinizing references.
Characteristics of studies included
A list of details abstracted from each study included in the meta-analysis is listed in . The studies in this meta-analysis, including 25 journal articles and 1 master’s thesis, were published from 2000 to 2013. Three of included studies had researched patients from different geographic areas, because different prevalence of DN, they were treated as independent studies.Citation30,Citation43,Citation50 Furthermore, one of included studies that had researched both T1DM patients and T2DM patients was treated as two independent studies.Citation38 Hence, the 31 references including 6144 cases and 4900 DM controls were included for our meta-analysis, 15 references including 2134 cases and 2348 non-DM controls were included. Among included studies, three studies had reported three allele (a, b and c allele) and four genotypes (b/b, b/a, a/a and b/c),Citation16,Citation30,Citation50 that distinguish from other studies. Our data analysis did not include c allele and b/c genotype, but the number of cases was invariable. According to “nine geographic race”, populations were categorized into Asian, Caucasian, African and Indian. Basing on skin color, populations were categorized into Asian, which is just the synthesis of Asian and Indian in “nine geographic race”, and Caucasian, African. So, the following analyses were conducted according to “nine geographic race” preferentially. Seventeen studies were on Asian population, 10 studies were about Caucasian population, 2 studies were about African population and 2 studies were about Indian population. Five studies focused on T1DM, and 26 studies paid attention to T2DM. When chose non-DM crowed as controls, there were not studies focused on T1DM patients and Indian population. The number of patients and controls, genotypes frequency and the test for HWE were also listed in .
Table 1. Characteristics of the studies included in the meta-analysis.
Table 2. Distribution of NOS3 genotypes and p values in χ2 test of HWE in the eligible studies.
Risk of bias assessment
The results of quality assessment using NOS for each study are listed in . As listed in , superior number of stars reflects the better quality, and the average number of stars in all studies was 4.73. There were only three studies to get 7 ∼ 9 stars (11.54%). Thirteen studies got one or zero for selection or zero for comparability or for exposure (50%), judging to have low quality or high risk of bias.
Table 3. Quality indicators from Newcastle-Ottawa scale (NOs).
Heterogeneity
When chose patients with DM as controls, there was a significant heterogeneity with respect to association between 4b/a polymorphism and DN under pre-allele model (p = 0.000, I2 = 57.7%) and dominant model (p = 0.000, I2 = 53.6%). In the subgroup analysis according to geographic race, there still existed heterogeneity for Asian population under pre-allele model (p = 0.000, I2 = 64.4%) and dominant model (p = 0.001, I2 = 57.7%). In the subgroup analysis according to type of DM, there also existed heterogeneity for T2DM under pre-allele model (p = 0.000, I2 = 59.8%) and dominant model (p = 0.001, I2 = 51.9%), for T1DM under dominant model (p = 0.016, I2 = 67.3%). In the subgroup analyses according to reporting the age (report and not report), BMI (report and not report), diabetes duration (> 10 years and ≤ 10 years), genotyping method (polymerase chain reaction-restricted fragment length polymorphisms and others), source of DM/DN (hospital, population and not report) and status of HWE in DM group (in HWE and not), there still existed heterogeneity under pre-allele model and dominant model in some degree. But I2 that testing the heterogeneity reduced to <50% in the subgroup diabetes duration >10 year under dominant model (p = 0.025, I2 = 48.7%), and in the subgroup “in HWE” under pre-allele model (p = 0.003, I2 = 48.8%) and dominant model (p = 0.007, I2 = 44.1%).
When chose populations without DM as controls, significant heterogeneity existed under pre-allele model (p = 0.000, I2 = 72.1%) and dominant model (p = 0.002, I2 = 58.4%). In the subgroup analysis according to ethnicity, there still existed heterogeneity for Asian population under pre-allele model (p = 0.000, I2 = 73.1%) and dominant model (p = 0.017, I2 = 55.5%), for Caucasian population under pre-allele model (p = 0.007, I2 = 75.0%) and dominant model (p = 0.028, I2 = 57.1%). The type of DM in studies using non-DM populations as controls was all T2DM, and all of them reported the age of DN patients. In the subgroup analyses according to BMI, diabetes duration, genotyping method and source of DM/DN, there also existing heterogeneity under pre-allele model and dominant model. In the subgroup analysis according to status of HWE in non-DM group, significant heterogeneity decreased under dominant model from total value p = 0.002, I2 = 58.4% to “in HWE” group p = 0.120, I2 = 32.8% and “not in HWE” group p = 0.181, I2 = 44.2%. The pooled and subgroups’ heterogeneity results based on geographic race, type of DM and HWE are listed in .
Table 4. Random effects odds ratios and heterogeneity results for the association of eNOS 4b/a gene polymorphisms and DN.
Association of the 4b/a polymorphism with DN and subgroup analyses (ethnicity analysis is geographic race analysis) when compared with DM patients
Because hypothesis of homogeneity was rejected by the I2 statistic, REM was used to evaluate the association of the 4b/a polymorphism with DN. Under the pre-allele model, the pooled OR suggested that NOS3 4a was associated with an increased risk of DN when compared with the 4b allele in global populations (OR = 1.26, 95% CI: 1.10–1.45) and in the Asian population (OR = 1.51, 95% CI: 1.13–2.01) (). Under the dominant model, 4aa + 4ab genotype was also associated with the risk of DN in global populations (OR = 1.27, 95% CI: 1.09–1.48) and in the Asian population (OR = 1.54, 95% CI: 1.16–2.05) when compared with the 4bb genotype (). Under the recessive model, in comparison with 4bb + 4ab genotype, 4aa genotype was significantly associated with the risk of DN in global populations (OR = 1.50, 95% CI: 1.12–2.02), Asian population (OR = 2.32, 95% CI: 1.30–4.15) and Indian population (OR = 5.32, 95% CI: 2.05–13.77) (). The rest additive model (4aa vs. 4bb) also produced similar associations (global populations, OR = 1.52, 95% CI: 1.13–2.04, Asian population OR = 2.38, 95% CI: 1.33–4.29) and Indian population (OR = 4.80, 95% CI: 1.84–12.49) (). As shown in , under the co-dominant genetic model, no association between 4ab genotype and the risk of DN, in comparison with 4aa + 4bb, was seen in global populations or any subgroups.
Figure 2. Meta-analysis for eNOS 4b/a polymorphism in DN (pre-allele model: 4a vs. 4b) compared with DM patients.
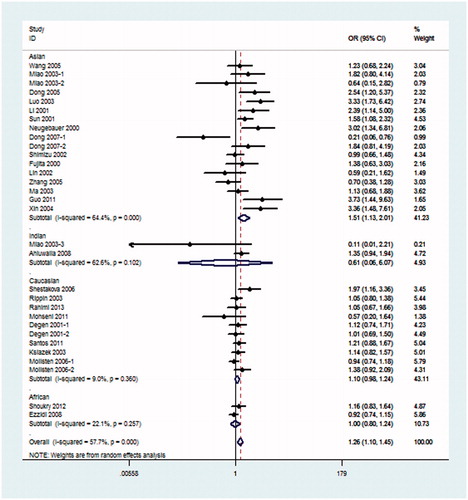
Figure 3. Meta-analysis for eNOS 4b/a polymorphism in DN (dominant model: 4aa + 4ab vs. 4bb) compared with DM patients.
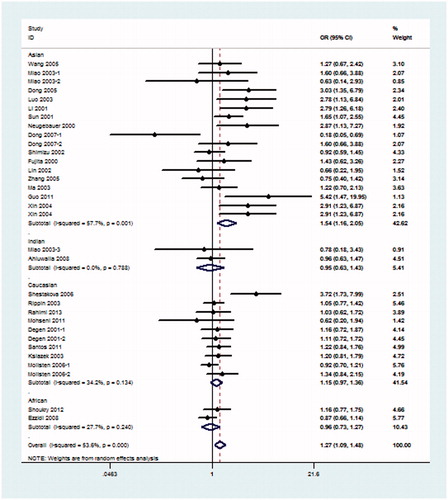
Figure 4. Meta-analysis for eNOS 4b/a polymorphism in DN (recessive model: 4aa vs. 4bb + 4ab) compared with DM patients.
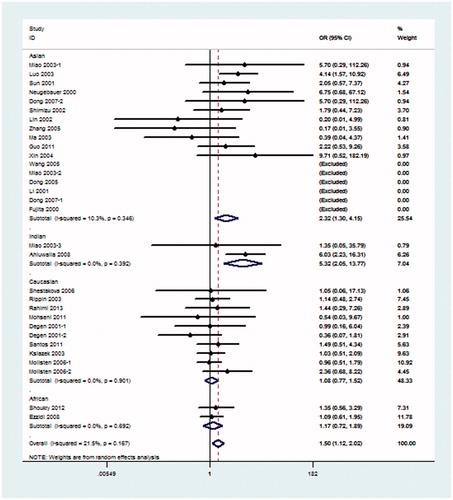
Figure 5. Meta-analysis for eNOS 4b/a polymorphism in DN (additive model: 4aa vs. 4bb) compared with DM patients.
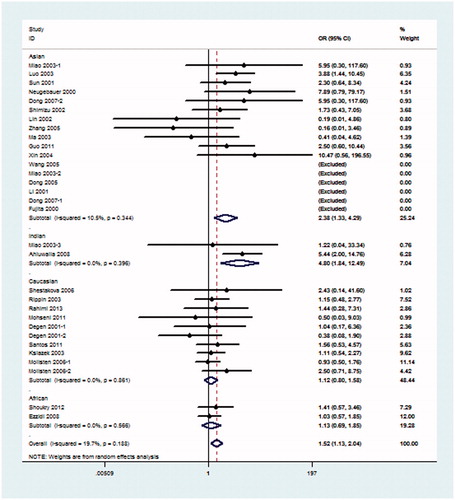
Figure 6. Meta-analysis for eNOS 4b/a polymorphism in DN (co-dominant model: 4ab vs. 4aa + 4bb) compared with DM patients.
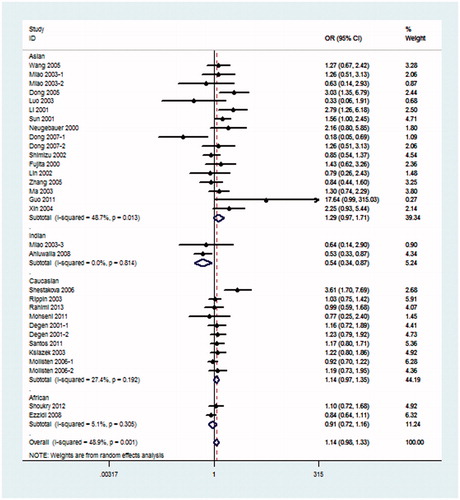
In the subgroup type of DM analyses, patients that had suffered from T2DM may have a risk of DN under the pre-allele model (OR = 1.29, 95% CI: 1.09–1.54), dominant model (OR = 1.28, 95% CI: 1.08–1.53), recessive model (OR = 1.65, 95% CI: 1.12–2.42) and additive model (OR = 1.66, 95% CI: 1.14–2.42). No association between DN patients that had suffered from T1DM and NOS3 4b/a was found ().
Association of the 4b/a polymorphism with DN and ethnicity analysis when compared with non-DM patients
Because I2 > 50% in the pre-allele model and dominant model, REM was fit for the meta-analysis. As listed in , there were obviously significant results between the 4b/a polymorphism and DN risk in the global populations under the recessive model (OR = 3.13, 95% CI: 1.55–6.32) and the additive model (OR = 3.41, 95% CI: 1.66–7.02), also the pre-allele model (OR = 1.74, 95% CI: 1.33–2.28), the dominant model (OR = 1.67, 95% CI: 1.30–2.16) and the co-dominant model (OR = 1.35, 95% CI: 1.15–1.60). In the Asian population, association was observed in the pre-allele model (OR = 2.09, 95% CI: 1.36–3.22), the dominant model (OR = 2.02, 95% CI: 1.39–2.93), the additive model (OR = 3.59, 95% CI: 1.06–12.14) and the co-dominant model (OR = 1.65, 95% CI: 1.28–2.11). No association between NOS3 4b/a and the risk of DN was found in the Caucasian population in any models. There was only one study including the African population (). Subgroup analysis for the type of DM was not conducted because the type of DM in studies using non-DM populations as controls was all T2DM.
Association of the 4b/a polymorphism with DN and ethnicity analysis according to skin color when compared with DM patients
In the ethnicity analysis according to skin color, there existed heterogeneity for Asian population under co-dominant model (p = 0.001, I2 = 57.7%), pre-allele model (p = 0.000, I2 = 62.4%) and dominant model (p = 0.001, I2 = 56.8%).
Because there still existed heterogeneity in the ethnicity analysis according to skin color, REM was used to evaluate the association of the 4b/a polymorphism with DN yet. In addition, when choosing skin color as subgroup, the disparate results in the Asian population were as follows, the pre-allele model: OR = 1.47, 95% CI: 1.13–1.91; the dominant model: OR = 1.46, 95% CI: 1.12–1.90; the recessive model: OR = 2.73, 95% CI: 1.61–4.62; the additive model: OR = 2.77, 95% CI: 1.66–4.62; the co-dominant genetic model: OR = 1.17, 95% CI: 0.88–1.57.
Sensitivity analyses
When chose DM patients as controls, under the FEM, sensitivity analyses for HWE (excluding studies controls were not in HWE) produced no significant changes in the pre-allele model (OR = 1.12, 95% CI: 1.03–1.22), dominant model (OR = 1.12, 95% CI: 1.02–1.23), recessive model (OR = 1.42, 95% CI: 1.10–1.83), additive model (OR = 1.41, 95% CI: 1.09–1.83) and co-dominant model (OR = 1.05, 95% CI: 0.95–1.17). Sensitivity analyses for diabetes duration (excluding studies DM duration ≤10 years) also yielded non-significant results. If we chose non-DM populations as controls, under the FEM, sensitivity analyses for HWE also did not derive significant changes (). Similar sensitivity analyses were also performed in comparisons of other subgroups, the pooled OR (including 95% CI) pattern was not changed consistently in all contrasts when each single study was excluded. Sensitivity analyses indicated the results of meta-analysis were stable.
Publication bias
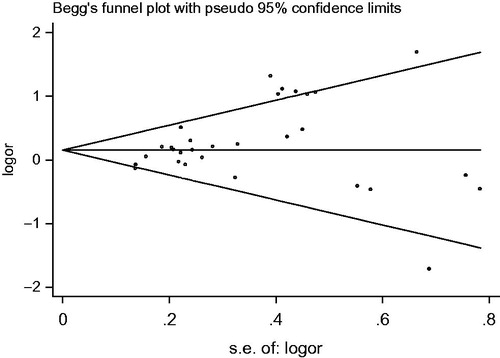
No study included in the meta-analysis reported that genotyping was performed blindly in clinical status. When chose DM patients as controls, under the dominant model, the shape of the funnel plot was asymmetrical and p value of Egger’s test was 0.024, indicating apparent publication bias (). But under other models, no significant publication bias was found. Funnel plots and Egger’s test for articles associated with NOS3 polymorphism and DN compared with non-DM populations indicated that there was no significant publication bias.
Discussion
It remains an unsolved problem why some diabetic patients develop DN, whereas others do not, despite both having a long-term hyperglycemia. Because known environmental factors did not fully explain this, researchers have explored the answer at the genetic level. There existed many experiment studies indicating significant association between NOS3 4b/a polymorphism and susceptibility for DN,Citation32–34 nevertheless other studies provided inverse results.Citation36,Citation45 A meta-analysis aiming at assessing overall effects of variants of the NOS3 4b/a on DN comparing with DM or non-DM was performed. In total, the meta-analysis involved 26 studies for DN comparing with DM and 15 studies for DN comparing with non-DM, which provided 6144/4900 cases/controls and 2134/2348 cases/controls, respectively. In the meta-analysis, the effects of the pre-allele, dominant, recessive, additive and co-dominant models were estimated. Moreover, the heterogeneity of genetic effects across ethnicity, type of DM, age, BMI, DM duration, genotyping method, source of DM/DN and status of HWE in controls were investigated. In addition, subgroup analyses based on ethnicity, the type of DM and sensitivity for excluding studies not in HWE were performed.
In this meta-analysis, when compared with DM, an association between NOS3 4b/a variants and DN risk was observed, especially in the Asian population and T2DM population. But because the most of the included studies is from Asia and the most of the included population is the patients of T2DM, the conclusion in global populations is not affirmative. When compared with non-DM crowed, an obvious association was found in the global populations and Asian population. According to these results, the effect size OR is lager when meta-analysis conducted between DN patients and non-DM populations, rather than DM patients. It is because patients with DM have higher risk to diabetic complications DN.Citation51 Maybe, the different endogenous environment of DM patients contributes to the pathology and development of DN. So, the DM patient without DN is a better control to test the effect of NOS3 polymorphisms and DN risk. According to above-mentioned results, we can conclude that the variant of NOS3 4b/a is a risk factor in the development and progress of DN in Asian population. This conclusion was consistent with published experiment researchesCitation34 and reviews,Citation52 also, was demonstrated in NOS3 knock-out mice model.Citation53 However, the testing of associations was based on different amount of information, moderate heterogeneity, the using of REM, different ethnicity division and low quality of evidence, Therefore, the results of our meta-analysis should be interpreted with caution.
Just as studies reported, endothelial dysfunction may give rise to uncoupling of the vascular endothelial growth factor (VEGF)-endothelial nitric oxide (eNO) axis, resulting in increased levels of VEGF and excessive proliferation of endothelial cell.Citation54,Citation55 Thus, maintaining the coupling of VEGF-eNO was deemed to have a crucial role in developing into DN.Citation56 The variant of NOS3 4b/a contributes to the decreased activity of NOS3 enzyme, which resulting in low basal levels of plasma NO, so, the level of VEGF increases, endothelial cell proliferates and develops into DN finally.Citation57 Furthermore, such hypothesis was proved in experimental mice model.Citation58 However, in our subgroup analysis, there was an increased risk among Asian population, but not other race. The reasons may as follows: (1) the distinguishable genetic background of pathology in the Asian population may contribute to such heterogeneity.Citation59 As listed in the , the frequency of every genotype was different significantly among different ethnicities: (1) It supported discrepancy of pooled OR statistically; (2) different linkage map, lifestyle, geographical environment or public sanitation may influence the activity of NOS3; (3) the criterion of ethnicity division is inappropriate; and (4) the CIs for ORs in different populations overlapped with each other, so, it is possible that the effect of NOS3 polymorphisms with the DN risk is similar among them. It is because the relevant researches from Indian (221/266 patients/controls), Caucasian (2463/2283 patients/controls) and African (705/598 patients/controls) population are statistically insufficient so as to cause such discrepancy. It was widely accepted that the existence of gene–environment interaction may result in the development of DN,Citation59 it can explain the discrepancy of results between the individual genetic association studies. In our subgroup analysis, there was an increased risk among T2DM. The reason for this result have not been explored by studies, one of probable reason is minor references in T1DM population.
The unadjusted pooled ORs were calculated in the meta-analysis, it is because that the possible confounding factors that affect the estimates of associations like age, sex, lifestyle, surroundings, physical condition (e.g. BMI, SBP/DBP, HbA1c, smoking, alcohol consumption and control of DM), exercise, duration, source of sampling and others were not provided clearly. Studies including in our meta-analysis exhibited moderate heterogeneity and the source of heterogeneity may derive from ethnicity, type of DM, diabetes duration and status of HWE. The existence of above multi-factors among studies may result in the presence of heterogeneity. For example, the prevalence of DN depends on age, it is easier to occur in elderly individuals. Thus, younger DM patients who may have suffered from DN were frequently included as controls. Therefore, if a control group includes cases that are still at risk of developing DN, the effect size may be minimized. It is the reason why we chose the overt DN as cases. At the same time, the strict selection criteria ensure the clear distinction in cases and controls of meta-analysis.
According to the funnel plot and Egger’s test, there was a significant publication bias under the dominant model when chose non-DM population as controls. On the basis of result of sensitivity analysis, omitting any single study will not change pooled results significantly. Thus, we think that the association between NOS3 polymorphisms and DN susceptibility will not be changed by publication bias. Our meta-analysis exist limitations inevitably because of following: (1) although we extracted characteristics from eligible studies and estimated their contributions to the heterogeneity separately, the combined effect of them can hardly be estimated. At the same time, we cannot take some environmental factors such as lifestyle, surroundings or exercises into account; (2) eligible studies are case–control studies which might have resulted in survival-related bias. Because DN patients carrying the risk allele may have the lower survival, and DN is a chronic disease, carriers of the risk genotype will be leakage of registration, so the power of effects may be reduced. Perspective study design of DM patients being followed up for the development of DN could solve this contradiction; (3) Asian study were maximum in our meta-analysis, the research from other race were minor, especially in Indian and African. In Asian subgroup, the sample size is minor generally, so the results from Asian population should be treated very carefully; (4) the quality of case–control study in our meta-analysis is low, that is high risk of bias. Furthermore, NOS was used to assess quality has not been agreed widely. Based on these bias and limitations, the any results calculated by our meta-analysis should be interpreted with caution.
In conclusion, this meta-analysis supports an increased susceptibility of NOS3 4b/a variant to DN in Asian population, the conclusion in global population needs more ethnic-specific association studies. Furthermore, to further demonstrate the role of NOS3 4b/a in the progress of DN, it is needed to carry out more perspective, high-quality studies in different populations with larger sample size and more matched clinical characteristics to avoid confounding factors, especially pay attention to the stage of DN and choose DM patients as controls. According to our meta-analysis, c allele and b/c genotype were reported in individual studies, so later studies should notice the gene detection.
Declaration of interest
The study did not accept any funding. There was not conflict of interest in our study.
References
- Ritz E, Rychlík I, Locatelli F, Halimi S. End-stage renal failure in type 2 diabetes: A medical catastrophe of worldwide dimensions. Am J Kidney Dis. 1999;34(5):795–808
- Newman DJ, Mattock MB, Dawnay ABS, et al. Systematic review on urine albumin testing for early detection of diabetic complications. Health Technol Assess. 2005;9(30):iii–vi, xiii–163
- Shpichinetsky V, Raz I, Friedlander Y, et al. The association between two common mutations C677T and A1298C in human methylenetetrahydrofolate reductase gene and the risk for diabetic nephropathy in type II diabetic patients. J Nutr. 2000;130(10):2493–2497
- Zintzaras E, Uhlig K, Koukoulis GN, Papathanasiou AA, Stefanidis I. Methylenetetrahydrofolate reductase gene polymorphism as a risk factor for diabetic nephropathy: A meta-analysis. J Hum Genet. 2007; 52(11):881–890
- Nakagawa T. Uncoupling of the VEGF-endothelial nitric oxide axis in diabetic nephropathy: An explanation for the paradoxical effects of VEGF in renal disease. Am J Physiol Renal Physiol. 2007;292(6):F1665–F1672
- Palm F, Teerlink T, Hansell P. Nitric oxide and kidney oxygenation. Curr Opin Nephrol Hypertens. 2009;18(1):68–73
- Noiri E, Satoh H, Taguchi J-I, et al. Association of eNOS Glu298Asp polymorphism with end-stage renal disease. Hypertension. 2002;40(4):535–540
- Moncada S, Higgs A. The L-arginine-nitric oxide pathway. N Engl J Med. 1993;329(27):2002–2012
- Star RA. Intrarenal localization of nitric oxide synthase isoforms and soluble guanylyl cyclase. Clin Exp Pharmacol Physiol. 1997;24(8):607–610
- Knowles RG, Moncada S. Nitric oxide synthases in mammals. Biochem J. 1994;298(Pt 2):249–258
- Alderton W, Cooper C, Knowles R. Nitric oxide synthases: Structure, function and inhibition. Biochem J. 2001;357:593–615
- Augeri AL, Tsongalis GJ, Van Heest JL, Maresh CM, Thompson PD, Pescatello LS. The endothelial nitric oxide synthase -786 T>C polymorphism and the exercise-induced blood pressure and nitric oxide responses among men with elevated blood pressure. Atherosclerosis. 2009;204(2):e28–e34
- Wolff B, Braun C, Schlter C, et al. Endothelial nitric oxide synthase Glu298→Asp polymorphism, carotid atherosclerosis and intima-media thickness in a general population sample. Clin Sci. 2005;109:475–481
- Sinici I, Atalar E, Kepez A, et al. Intron 4 VNTR polymorphism of eNOS gene is protective for cardiac syndrome X. J Investig Med. 2010;58(1):23–27
- Rahimi Z, Shahvaisi-Zadeh F, Sadeghei S, Vessal M, Yavari N. eNOS 4a/b polymorphism and its interaction with eNOS G894T variants in type 2 diabetes mellitus: Modifying the risk of diabetic nephropathy. Dis Markers. 2013;34(6):437–443
- Santos KG, Crispim D, Canani LH, Ferrugem PT, Gross JL, Roisenberg I. Association of eNOS gene polymorphisms with renal disease in Caucasians with type 2 diabetes. Diabetes Res Clin Pract. 2011;91(3):353–362
- Ezzidi I, Mtiraoui N, Mohamed MB, Mahjoub T, Kacem M, Almawi WY. Association of endothelial nitric oxide synthase Glu298Asp, 4b/a, and -786T>C gene variants with diabetic nephropathy. J Diabetes Complications. 2008;22(5):331–338
- Neugebauer S, Baba T, Watanabe T. Association of the nitric oxide synthase gene polymorphism with an increased risk for progression to diabetic nephropathy in type 2 diabetes. Diabetes. 2000;49(3):500–503
- Definition, W. H. O. Part 1: Diagnosis and Classification of Diabetes Mellitus. Definition, Diagnosis and Classification of Diabetes Mellzitus and its Complications. Geneva: World Health Organization, Department of Noncommunicable Disease Surveillance; 1999
- Xue J, Chen LZ, Xue L, Zhou Q. Meta-analysis of risk factors for childhood cerebral palsy during pregnancy. CJCP. 2013;15(7):535–540
- Deeks JJ, Dinnes J, D’Amico R, et al. Evaluating non-randomised intervention studies. Health Technol Assess. 2003;7(27):1–173
- Wells G, Shea B, O’Connell D, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomized studies in meta-analyses. 2010. Available at: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp. Accessed July 2014
- Yang YL, Liu L, Wang Y, et al. The prevalence of depression and anxiety among Chinese adults with cancer: A systematic review and meta-analysis. BMC Cancer. 2013;13:393
- University of Thessaly, School of Medicine Laboratory of Biomathematics. Revealing the Mode of Inheritance in Genetic Association Studies. Available at: http://www.doc88.com/p-579886058890.html. Accessed April 2014
- Borenstein M, Hedges LV, Higgins JP, Rothstein HR. Part 3: Fixed-Effect Versus Random-Effects Models. Introduction to Meta-Analysis. Englewood: Wiley; 2011
- Alderson P, Creen S, Higgins JPT. Cochrane Reviewer’s Handbook4.2.2 (update MSTVH2004). Cochrane Library. Issue 1. Chechester: Wiley; 2004
- Garn SM. Human Races. Illinois: Thomas; 1971
- Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634
- Wang SL. Study of the Susceptibility Genes of Type 2 Diabetic Nephropathy in Northern Chinese. Shandong: Shandong University; 2005
- Miao L. Association of Nitric Oxide Synthase Gene Polymorphism with Diabetic Nephropathy in Type 2 Diabetic Patients. Shandong: Shandong University; 2003
- Dong YH, Qu SP, Lü WS, et al. Gene polymorphism in chromosome 7q35 and susceptibility to diabetic nephropathy. Chin J Diabetes. 2005;21(1):47–50
- Luo H, Ning YY. Association of polymorphism of endothelialnitricoxide synthase gene with diabetic nephropathy. Chin J Diabetes. 2003;11(5):317–320
- Shestakova M, Vikulova O, Gorashko N, et al. The relationship between genetic and hemodynamic factors in diabetic nephropathy (DN): Case–control study in type 1 diabetes mellitus (T1DM). Diabetes Res Clin Pract. 2006;74(2):S41–S50
- Li C, Dong Y, Lü W. [The association between polymorphism of endothelial nitric oxide synthase gene and diabetic nephropathy]. Zhonghua nei ke za zhi [Chin J Intern Med]. 2001;40(11):729–732
- Rippin J, Patel A, Belyaev N, Gill G, Barnett A, Bain S. Nitric oxide synthase gene polymorphisms and diabetic nephropathy. Diabetologia. 2003;46(3):426–428
- Mehrab-Mohseni M, Tabatabaei-Malazy O, Hasani-Ranjbar S, et al. Endothelial nitric oxide synthase VNTR (intron 4 a/b) polymorphism association with type 2 diabetes and its chronic complications. Diabetes Res Clin Pract. 2011;91(3):348–352
- Shoukry A, Shalaby SM, Abdelazim S, et al. Endothelial nitric oxide synthase gene polymorphisms and the risk of diabetic nephropathy in type 2 diabetes mellitus. Genet Test Mol Biomarkers. 2012;16(6):574–579
- Degen B, Schmidt S, Ritz E. A polymorphism in the gene for the endothelial nitric oxide synthase and diabetic nephropathy. Nephrol Dial Transplant. 2001;16(1):185–198
- Shimizu T, Onuma T, Kawamori R, Makita Y, Tomino Y. Endothelial nitric oxide synthase gene and the development of diabetic nephropathy. Diabetes Res Clin Pract. 2002;58(3):179–185
- Ahluwalia TS, Ahuja M, Rai TS, et al. Endothelial nitric oxide synthase gene haplotypes and diabetic nephropathy among Asian Indians. Mol Cell Biochem. 2008;314(1–2):9–17
- Ksiazek P, Wojewoda P, Muc K, Buraczynska M. Endothelial nitric oxide synthase gene intron 4 polymorphism in type 2 diabetes mellitus. Mol Diagn. 2003;7(2):119–123
- Sun H-Y, Yang M, Liu S, Wang C, Zhang Q, Chen M. Study on the correlation of the polymorphisms of endothelial nitric oxide synthase gene with type 2 diabetes mellitus and diabetic nephropathy. Chin J Prev Contr Chron Non-Commun Dis. 2004;12(3):101–103
- Möllsten A, Wessman M, Svensson M, et al. Glu298Asp and NOS4ab polymorphisms in diabetic nephropathy. Ann Med. 2006;38(7):522–528
- Fujita H, Narita T, Meguro H, et al. Lack of association between an ecNOS gene polymorphism and diabetic nephropathy in type 2 diabetic patients with proliferative diabetic retinopathy. Horm Metab Res. 2000;32(2):80–83
- Lin S, Qu HQ, Qiu MC. Study on the association between ecNOS4b/a polymorphism and diabetic nephropathy. Chin J Nephrol. 2002;18(4):258–261
- Zhang M, Liu H, Yang HY, Wang YM, Song DP, Li H. Relationship of endothelial nitric oxide synthase (eNOS) 4a/b gene polymorphism with diabetic nephropathy. Chin J Diabetes. 2005;11(4):284–285
- Ma JR. Study on the Relationship between the Endothelial Constitutive Nitric Oxide Synthase (ecNOS) Gene Polymorphism and the Development of Diabetic Vascular Complication in Type 2 Diabetic Patients. Tianjin: Tianjin Medical University; 2003
- Guo XJ, Liu SJ. The association between endothelial nitric oxide synthase gene polymorphism and type 2 diabetic nephropathy. China Healthc Innovat. 2012;6(19):3–5
- Xing Q, Su BL, Li CC, et al. Association between endothelial nitric oxide synthase gene polymorphism and type 2 diabetic nephropathy. Chin J Endocrinol Metab. 2004;20(5):435–437
- Dong J, Zhao J, Lee K, Lir M, Chan S, Liao L. [Distribution of polymorphism in endothelial nitric oxide synthase gene in Singapore Chinese and its association with diabetic nephropathy]. Zhonghua yi xue za zhi. 2007;87(48):3415–3417
- Turner R, Holman R, Cull C, et al. Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (UKPDS 33). Lancet. 1998;352(9131):837–853
- He Y, Fan Z, Zhang J, et al. Polymorphisms of eNOS gene are associated with diabetic nephropathy: A meta-analysis. Mutagenesis. 2011;26(2):339–349
- Nakagawa T, Sato W, Glushakova O, et al. Diabetic endothelial nitric oxide synthase knockout mice develop advanced diabetic nephropathy. J Am Soc Nephrol. 2007;18(2):539–550
- Masuda Y, Shimizu A, Mori T, et al. Vascular endothelial growth factor enhances glomerular capillary repair and accelerates resolution of experimentally induced glomerulonephritis. Am J Pathol. 2001;159(2):599–608
- Ziche M, Morbidelli L, Choudhuri R, et al. Nitric oxide synthase lies downstream from vascular endothelial growth factor-induced but not basic fibroblast growth factor-induced angiogenesis. J Clin Invest. 1997;99(11):2625–2634
- Brodsky SV, Gao S, Li H, Goligorsky MS. Hyperglycemic switch from mitochondrial nitric oxide to superoxide production in endothelial cells. Am J Physiol Heart Circ Physiol. 2002;283(5):H2130–H2139
- Zheng X, Lou J, Yang W. Role of nitric oxide on diabetic nephropathy. Chinese J Nephrol. 1999;2:1–113
- Hossain P, Kawar B, El Nahas M. Obesity and diabetes in the developing world—A growing challenge. N Eng J Med. 2007;356(3):213–215
- Liu M. Susceptibility Genes, Environmental Risk Factors and their Interactions in the Development of Type 2 Diabetes Mellitus. Wuhan:Huazhong University of Science and Technology; 2006