Abstract
To reevaluate the association between the costimulatory molecule cytotoxic T lymphocyte-associated antigen4 (CTLA4) single nucleotide polymorphism (SNP) +49A/G and acute rejection (AR) in renal transplantation, nine studies published before June 2013 were analyzed. Meta-analysis and cumulative meta-analysis (metacum) were performed for each genotype in a random/fixed effect model. The combined odds ratios (OR) with 95% confidence intervals (CI) were calculated to estimate the strength of the association. In the sensitivity analysis, a single study involved in the meta-analysis was deleted each time to investigate the influence of the individual data sets on the pooled ORs. Meta-analysis regression was used for some influence factors, such as year of publication, total number in each group (AR group and control group), ethnicity, the ratio of GG to GA + AA, the ratio of G to A in CTLA4 +49A/G. Overall, a significant correlation was noted between the CTLA4 SNP (+49A/G) and the risk of AR (for GG vs. AG + AA: OR = 1.35, 95% CI = 1.05–1.73, p = 0.02; for G vs. A: OR = 1.21, 95% CI = 1.03–1.42, p = 0.02), especially in the Asian subgroup (for GG vs. AG + AA: OR = 1.79, 95% CI = 1.15–2.78, p = 0.009; for G vs. A: OR = 1.47, 95% CI = 1.04–2.07, p = 0.03). Of the influence factors, the ratio of GG to GA+AA (p = 0.046) and the ratio of G to A (p = 0.017) were significant factors. In conclusion, our results suggest that CTLA4 +49A/G contribute to the risk of AR following renal transplantation.
Introduction
Cytotoxic T lymphocyte-associated antigen 4 (CTLA4) is a key element in the immune system that induces immune tolerance and is one of the critical negative regulators of the T cell-mediated immune response.Citation1 It is also expressed constitutively on the surface of regulatory T cells (Tregs) and is detectable on approximately 50% of Tregs; it is only found on <1% of naive helper T cells.Citation2 CTLA4 ligation on Tregs results in a significant decrease in the presentation capacity of the antigen-presenting cells and effector T cell downregulation in mice.Citation3 CTLA4 plays an important role in the downregulation of the immune response. The rs231775 (+49A/G) single nucleotide polymorphism (SNP) is located within the signal peptide of the molecule and influences the expression of the full-length isoform on the T cell membrane. The expression pattern of the CTLA4 protein is also changed by polymorphisms of the rs4553808 (−1661A/G) and rs5742909 (−318C/T) loci, which are located in the CTLA4 gene promoter.Citation4 Similarly, the rs733618 (−1772T) allele was found to decrease transcription of the CTLA4 gene by influencing the binding of transcription factors.Citation5 The rs3087243 (+6230G/A) SNP is situated within the 3′ untranslated region of the CTLA4 gene and was found to be associated with susceptibility to autoimmune diseases.Citation6 The +49A/G (rs231775) and the +6230G/A (rs3087243) SNPs of the CTLA4 gene play influential roles in graft rejection and the long-term clinical outcome of organ transplantation.Citation7–12 Among these polymorphisms, the +49A/G (rs231775) polymorphism is the most widely investigated in renal transplantation. Over the last few years, numerous studies have been conducted concerning the relationship between the CTLA4 +49A/G polymorphism, found in exon 1, and acute rejection (AR) following renal transplantation indifferent races and ethnicities. However, these studies have yielded inconsistent results.Citation13,Citation14 To produce more precise results, we evaluated these associations using a meta-analysis.
Materials and methods
Identification of eligible studies
The relevant literature was extracted from databases including MEDLINE and EMBASE; the last updated search was performed before July 2013. The following search terms were used: “CTLA4”, “polymorphism”, “kidney”, and “AR”. Only published articles were included in this study. The references of the selected papers were also manually checked for other relevant articles that might have been missed in the initial search. No limitation was set on the language of the literature. The following inclusion criteria were used: (1) the study discussed the association between the SNP CTLA4 +49A/G and the risk of AR; (2) the study described useful genotype frequencies; and (3) when a study reported results indifferent subpopulations, these subpopulations were considered as separate studies. The following exclusion criteria were used: (1) the study was conducted on animals; (2) there was no control group; (3) the study was not associated with the polymorphism; and (4) for studies with overlapping or repeated data, the most recent or complete studies with the largest numbers of cases and controls were included.
Data extraction
Two investigators (Yifeng Guo and Fang Guo) independently reviewed and extracted the information from all eligible publications according to the inclusion and exclusion criteria listed above. In the case of a conflict, an agreement was reached after a discussion between the two reviewers. The following characteristics were extracted from each study: name of the first author, year of publication, country of origin, racial descent of participants, polymorphisms, number of cases and controls, p value for HWE, and genotyping methods.
Statistical analysis
The allele frequencies of the CTLA4 gene polymorphisms were determined using the allele counting method. Hardy–Weinberg equilibrium (HWE) was assessed in each study using the goodness-of-fittest (chi-square test or Fisher exact test) in case-control groups. The pooled odds ratios (ORs) with 95% confidence intervals (95% CI) were used to assess the strength of the association. Analysis of the association between the CTLA4 polymorphism and AR was performed using a dominant model, a recessive model, a co-dominant model, and an allele model. Statistical heterogeneity among the studies was assessed using a chi-square test; a corresponding p value below 0.05 was considered to represent significant heterogeneity. Meta-regression analysis and metan-based influence analysis were performed to analyze the heterogeneity more deeply. If there was a significant difference in terms of heterogeneity, the ORs were pooled according to the random effect model (the Der Simonian and Laird model). Otherwise, a fixed effect model (the Mantel–Haenszel model) was used. Subgroup analysis was performed based on race. To assess the publication bias, the Egger’s regression test and the Begg–Mazumdar test based on Kendall'stau were carried out. Cumulative meta-analysis was performed by year of publication. All the statistical analyses were performed using Review Manerge 5.0 (Cochrane Collaboration, Oxford, UK) and Stata 12.0 (StataCorp LP, College Station, TX).
Results
Study characteristics and eligible studies
In total, 43 papers were identified after an initial search. After screening the articles (), nine of these articles were included. All nine studies () contained data for the rs231775 (+49G/A) polymorphism. When categorized by ethnicity, the subjects of six studies were European,Citation15–20 two were Asian,Citation21,Citation22 and the remaining subjects were African.Citation7 The characteristics of these studies are shown in .
Table 1. Characteristics of the studies included in the meta-analysis.
The rs231775 A/G (+49A/G) polymorphism
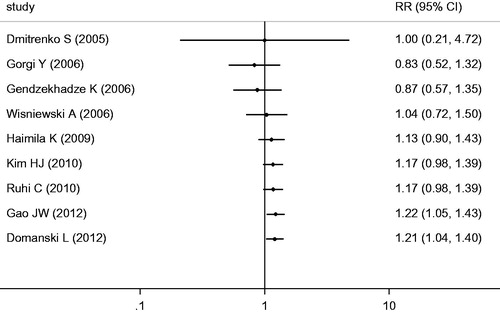
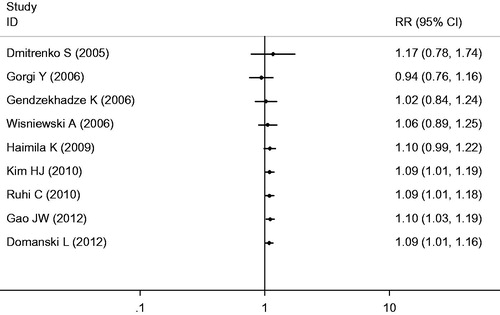
The eligible studies for the analysis of the +49A/G polymorphism included 481 cases with AR and 1324 non-AR controls. Overall, nine case-control studies were included in the meta-analysis of the association between the +49A/G polymorphism and the risk of AR. Among these case-control studies, six were from Europe, two were from Asia, and one was from Africa. While a significant association was observed for the GG versus AG + AA genotype (OR = 1.35, 95% CI = 1.05–1.73, p = 0.02) and the G versus A allele (OR = 1.21, 95% CI = 1.03–1.42, p = 0.02), the comparisons of the other genotypes did not reveal any statistical association (, ). In the subgroup meta-analysis, a significant association was observed in the analysis of the GG versus AG + AA genotype (OR = 1.79, 95% CI = 1.15–2.78, p = 0.009) and the G versus A allele (OR = 1.47, 95% CI = 1.04–2.07, p = 0.03) genotype in the Asian group (, and ).
Figure 2. Meta-analysis for the association between AR risk in renal transplantation and CTLA4 +49A/G (GG + AG vs. AA).
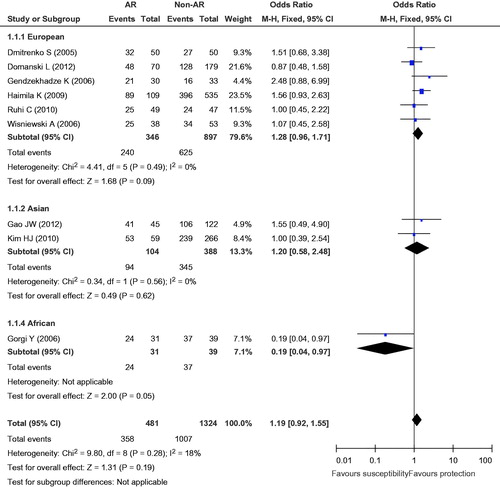
Figure 3. Meta-analysis for the association between AR risk in renal transplantation and CTLA4 +49A/G (GG vs. AG + AA).
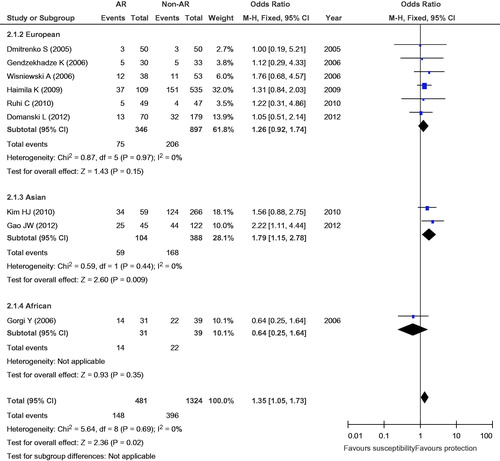
Figure 4. Meta-analysis for the association between AR risk in renal transplantation and CTLA4 +49A/G (GG vs. AA).
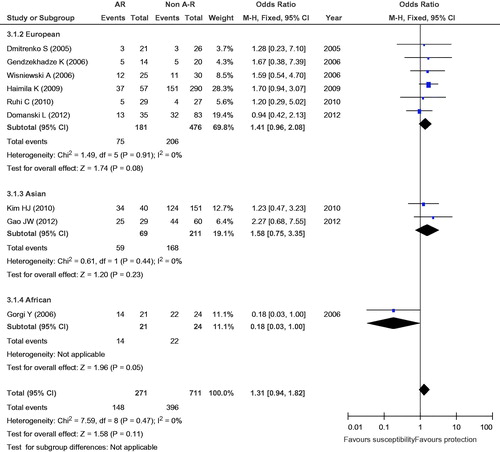
Figure 5. Meta-analysis for the association between AR risk in renal transplantation and the CTLA4 +49A/G (AG vs. AA).
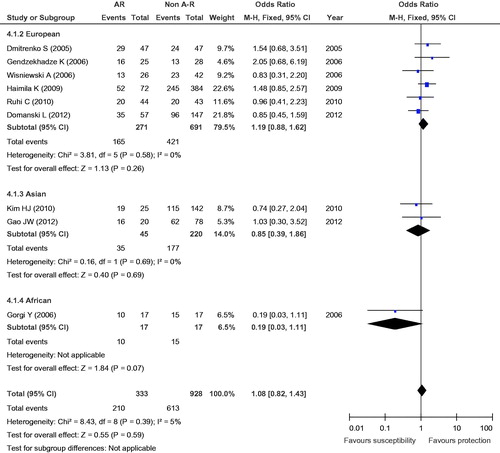
Figure 6. Meta-analysis for the association between AR risk in renal transplantation and CTLA4 +49A/G (G vs. A).
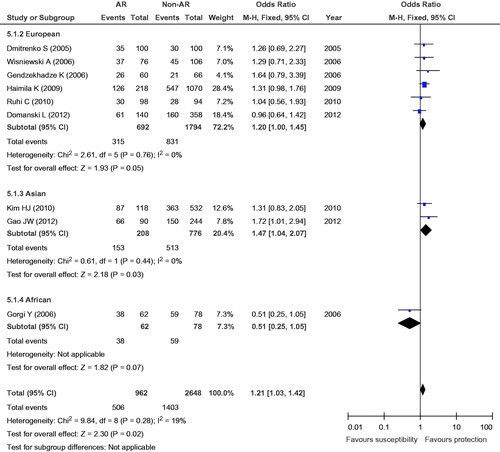
Table 2. Results from different comparative genetic models in CTLA4 +49A/G (rs231775).
Sensitivity analysis
A single study involved in the meta-analysis was deleted each time to investigate the influence of the individual data sets on the pooled ORs. As expected, the results of the +49A/G analysis were sensitive because of the marginal p value. After excluding one study, we identified a statistically significant change in the OR (95% CI). For the GG versus AG + AA genotype, after removing the Gorgi et al.Citation7 study, the p value changed from 0.02 to 0.007; after removing the Kim et al.Citation21 study, p = 0.02 changed to p = 0.06. Removal of the Gao et al.Citation22 study, p = 0.02 changed to p = 0.10. For the GG versus AA genotype, the removal of the Gorgi et al.Citation7 study, the p value changed from 0.11 to 0.03. For the G versus A allele, after removing the Gorgi et al.Citation7 study, the p value changed from 0.02 to 0.006; removal of the Domanski et al.Citation20 study, p = 0.02 changed to p = 0.009, and the removal of the Gao et al.Citation22 study, the p value changed from 0.02 to 0.08. Other corresponding pooled ORs were not significantly altered (data not shown).
Heterogeneity analysis
To assess the stability of the results of the meta-analysis, statistical heterogeneity among the studies was assessed using a chi-square test; a corresponding p value below 0.05 was considered to represent significant heterogeneity. As shown in , all the results did not show significant heterogeneity (GG + GA vs. AA: I2 = 18%, p = 0.28; GG vs. GA + AA: I2 = 0, p = 0.69; GG vs. AA: I2 = 0, p = 0.47; GG vs. GA: I2 = 5%, p = 0.39; G vs. A: I2 = 19%, p = 0.28).
Meta-analysis regression
To model the GG versus GA + AA and the G versus A effects, meta reg analysis was used for some influence factors, such as year of publication, total number in each group (such as the AR group and control group), ethnicity, the ratio of GG to GA + AA, or the ratio of G to A for the CTLA4 +49A/G SNP. Of these factors, the ratio of GG to GA + AA (p = 0.046) and the ratio of G to A (p = 0.017) were significant.
Cumulative meta-analysis
Cumulative meta-analysis was used based on the year of publication (from 2005 to 2012) to assess the CTLA4 +49GG versus GA + AA and the G versus A genotypes ( and ). OR point estimates and CI stabilized, and there was a good change in the trend situation.
Publication bias
Publication bias was assessed using Begg’s test and Egger’s test. Funnel plot asymmetry was assessed using Egger’s linear regression test. If the line passed through the origin, this indicated that publication bias did not exist. Excluding the +49GG + AG versus AA (Begg’s test, p = 0.009; Egger’s test, p = 0.021) and +49GG versus AA (Begg’s test, p = 0.048; Egger’s test, p = 0.081) comparisons, Begg’s test and Egger’s test suggested no publication bias ().
Discussion
The CTLA4 gene has been widely studied, and many studies have evaluated the effect of polymorphisms in the CTLA4 gene on AR in transplant recipients. Some studies have yielded conflicting results. Duan and Zhu performed a meta-analysis to evaluate these associations in AR following renal and liver transplantation, respectively.Citation13,Citation14 In Duan’s study,Citation13 the association between the CTLA4 +49G allele and AR was weakly significant (OR = 0.805, 95% CI = 0.677–0.957, p = 0.014), but in the meta-analysis from Zhu,Citation14 no significant association was discovered between the +49G/A SNP and AR in kidney transplantation. To produce more precise results, we enlarged the number of studies used in the meta-analysis to include a total of nine articles published by 2013; among these papers, six were from Europe, two were from Asia, and one was from Africa. For the +49A/G SNP, no significant results have been discovered from the present statistical data from a single study. Only in Korean patientsCitation21 was the CTLA4 +49A/G SNP (rs231775) statistically associated with late acute rejection (LAR) in the dominant model (OR = 0.48, 95% CI = 0.25–0.93, corrected p = 0.026); the allele frequency of the same SNP (rs231775) was also statistically associated with a risk of LAR (OR = 2.02, 95% CI = 1.15–3.52, corrected p = 0.013), where the presence of the G allele increased the risk of LAR in kidney transplantation. However, meta-analysis of the renal transplantation data shows that recipients carrying the GG genotype and the G allele had an increased risk of AR (GG vs. GA + AA and G vs. A; p = 0.02).
The metareg analysis was used for some influencing factors, such as year of publication, total number in each group (AR group and control group), ethnicity, the ratio of GG to GA + AA, and the ratio of G to A alleles. Of these factors, we discovered that there were statistically significant differences between studies for the ratio of GG to GA + AA (p = 0.046) and the ratio of G to A (p = 0.017). In a meta-analysis, examining the association of the CTLA4 gene with Graves’ disease in the Chinese Han population in the large samples, the ratio of the G and A alleles was 70.5/29.5 = 2.39 in Chinese populations (healthy controls).Citation23 In meta-analyses, examining the association between the CTLA4 exon1 +49A/G polymorphism and systemic lupus erythematosusCitation24 in the Korean population, the ratio of G to A was 1.950 (Pyo et al.Citation25), 2.053 (Hudson et al.Citation5), and 2.822 (Lee et al.Citation26); in the Japanese population, the G/A ratio was 1.339 (Ahmed et al.Citation27), 1.586 (Matsushita et al.Citation28), and 2.014 (Takeuchi et al.Citation29). The Spanish population had a G/A ratio of 0.352 (Aguilar et al.Citation30), whereas the Portuguese population had one of 0.378 (Barreto et al.Citation31), and the English population was 0.458 (Heward et al.Citation32). In another paper, the G/A ratio was 0.876 in the Italian population (healthy controls) (Brozzetti et al.Citation33). In other words, the MAF (minor allele frequency) of the G and A alleles at the +49 locus of the CTLA4 gene may be different between Asian and European populations; the minor allele may be A in Asia, but G in Europe.
The subgroup analysis (GG vs. GA + AA and G vs. A) in the Asian subgroup, there were significant results (GG vs. GA + AA, p = 0.009; G vs. A, p = 0.03), but there were no statistically significant results in the European subgroup (GG vs. GA + AA, p = 0.15; G vs. A, p = 0.05). In the sensitivity analysis, when the Asian studies were removed one by one the statistics became meaningless. Therefore, the ethnicity and the ratio of the G and A alleles may be internal factors that influence the development of AR in renal transplant recipients.
The cell-surface expression of CTLA4 was significantly increased in individuals carrying the AA genotype compared to the expression in carriers of the AG and GG genotypes.Citation34 T cells with the +49GG genotype had higher activation and proliferation rates compared to those with the +49AA genotype.Citation35 CTLA4 +49G > A caused a 17Ala > 17Thr substitution in the leading peptide of CTLA4.Citation36 The 17Thr substitution increased the binding of CTLA4 to B7.1, causing stronger inhibition of T cell activation than CTLA4 17Ala.Citation35 Recently, the G allele of the +49A/G polymorphism was reported to have a strong association with autoimmune diseases.37–39 In our paper, the G allele was associated with AR in kidney transplantation; there may be a similar immune mechanism involved in autoimmune diseases.
Heterogeneity and publication bias can influence the results of meta-analyses. There was no significant heterogeneity in the overall comparisons for all five polymorphisms. Therefore, heterogeneity did not appear to influence the results, suggesting that our results were reliable. In our meta-analysis, only studies indexed by the selected databases were included. Negative studies were less likely to be published in journals or to be available in computerized databases, resulting in potential overestimation of effect sizes.40 In this meta-analysis, Begg’s test and Egger’s test showed significant publication bias in the model for GG versus AG + AA and GG versus AA, so the current results should be interpreted cautiously. The limitations of this meta-analysis should be considered. First, the number of available studies that could be included was relatively small. Second, only two of the nine studies were conducted in Asian populations, and only one of the nine studies was conducted in an African population. Third, the overall outcomes were based on individual unadjusted ORs. Fourth, there was a lack of a general allele survey of the CTLA4 +49A/G locus in the various populations, and the inconsistency of the minor allele of A/G between the European and Asian populations needs to be confirmed.
Overall, the current meta-analysis suggests that the +49A/G polymorphism in the CTLA4 gene may be associated with the risk of rejection after renal transplantation, especially in the Asian population. Well-designed, unbiased prospective studies with larger sample sizes that address the gene–ethnicity interactions should be conducted to confirm these results.
Declaration of interest
The authors report no conflicts of interest.
The project was partly supported by grants from the Chinese Postdoctoral Science Foundation (2003033271).
References
- Alegre ML, Frauwirth KA, Thompson CB. T-cell regulation by CD28 and CTLA-4. Nat Rev Immunol. 2001;1(3):220–228
- Grohmann U, Orabona C, Fallarino F, et al. CTLA-4-Ig regulates tryptophan catabolism in vivo. Nat Immunol. 2002;3(11):1097–1101
- Wing K, Onishi Y, Prieto-Martin P, et al. CTLA-4 control over Foxp3+ regulatory T cell function. Science. 2008;322(5899):271–275
- Bouqbis L, Izaabel H, Akhayat O, et al. Association of the CTLA4 promoter region (−1661G allele) with type 1 diabetes in the South Moroccan population. Genes Immun. 2003;4(2):132–137
- Hudson LL, Rocca K, Song YW, Pandey JP. CTLA-4 gene polymorphisms in systemic lupus erythematosus: A highly significant association with a determinant in the promoter region. Hum Genet. 2002;111(4–5):452–455
- Ueda H, Howson JM, Esposito L, et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423(6939):506–511
- Gorgi Y, Sfar I, Abdallah TB, et al. Ctla-4 exon 1 (+49) and promoter (−318) gene polymorphisms in kidney transplantation. Transplant Proc. 2006;38(7):2303–2305
- Wu J, Tang JL, Wu SJ, Lio HY, Yang YC. Functional polymorphism of CTLA-4 and ICOS genes in allogeneic hematopoietic stem cell transplantation. Clin Chim Acta. 2009;403(1–2):229–233
- Kusztal M, Kościelska-Kasprzak K, Drulis-Fajdasz D, et al. The influence of CTLA-4 gene polymorphism on long-term kidney allograft function in Caucasian recipients. Transpl Immunol. 2010;23(3):121–124
- Slavcheva E, Albanis E, Jiao Q, et al. Cytotoxic T-lymphocyte antigen 4 gene polymorphisms and susceptibility to acute allograft rejection. Transplantation. 2001;72(5):935–940
- Tapirdamaz O, Pravica V, Metselaar HJ, et al. Polymorphisms in the T cell regulatory gene cytotoxic T lymphocyte antigen 4 influence the rate of acute rejection after liver transplantation. Gut. 2006;55(6):863–868
- deReuver P, Pravica V, Hop W, et al. Recipient ctla-4 +49G/G genotype is associated with reduced incidence of acute rejection after liver transplantation. Am J Transplant. 2003;3(12):1587–1594
- Duan Z, Zhang Y, Pan F, et al. Association between CTLA4 gene polymorphisms and acute rejection of kidney transplantation: A meta-analysis. J Nephrol. 2012;25(6):996–1002
- Zhu CL, Huang Q, Liu CH, Xie F. Polymorphisms in the cytotoxic T-lymphocyte antigen 4 gene and acute rejection risk in transplant recipients. Mol Biol Rep. 2012;39(9):8701–8708
- Dmitrienko S, Hoar DI, Balshaw R, Keown PA. Immune response gene polymorphisms in renal transplant recipients. Transplantation. 2005;80(12):1773–1782
- Wisniewski A, Kusztal M, Magott-Procelewska M, et al. Possible association of cytotoxic T-lymphocyte antigen 4 gene promoter single nucleotide polymorphism with acute rejection of allogeneic kidney transplant. Transpl Proc. 2006;38(1):56–58
- Gendzekhadze K, Rivas-Vetencourt P, Montano RF. Risk of adverse post-transplant events after kidney allograft transplantation as predicted by CTLA-4 +49 and TNF-a-308 single nucleotide polymorphisms: A preliminary study. Transpl Immunol. 2006;16(3–4):194–199
- Haimila K, Turpeinen H, Alakulppi NS, Kyllönen LE, Salmela KT, Partanen J. Association of genetic variation in inducible costimulator gene with outcome of kidney transplantation. Transplantation. 2009;87(3):393–396
- Ruhi C, Sallakci N, Ersoy F, Yegin O, Süleymanlar G. The relation between CTLA-4 single nucleotide polymorphisms and acute rejection in kidney transplantation. NDT Plus. 2010;Suppl 3:iii524–iii525
- Domański L, Bobrek-Lesiakowska K, Kłoda K, et al. The impact of rs231775 (+49AG) CTLA4 gene polymorphism on transplanted kidney function. Ann Transplant. 2012;17(3):29–35
- Kim HJ, Jeong KH, Lee SH, et al. Polymorphisms of the CTLA4 gene and kidney transplant rejection in Korean patients. Transpl Immunol. 2010;24(1):40–44
- Gao JW, Guo YF, Fan Y, et al. Polymorphisms in cytotoxic T lymphocyte associated antigen-4 influence the rate of acute rejection after renal transplantation in 167 Chinese recipients. Transpl Immunol. 2012;26(4):207–211
- Zhao SX, Pan CM, Cao HM, et al. Association of the CTLA4 gene with Graves’ disease in the Chinese Han population. PLoS One. 2010;5(3):e9821
- Chang WW, Zhang L, Yao YS, Su H. Association between CTLA-4 exon-1 +49A/G polymorphism and systemic lupus erythematosus: An updated analysis. Mol Biol Rep. 2012;39(9):9159–9165
- Cho CS, Kim HY, Pyo CW, et al. The distribution of CTLA-4 alleles in Korean patients with SLE. Korean J Immunol. 2000;22:17–22
- Lee YH, Kim YR, Ji JD, Sohn J, Song GG. Polymorphisms of the CTLA-4 exon 1 and promoter gene in systemic lupus erythematosus. Lupus. 2001;10(9):601–605
- Ahmed S, Ihara K, Kanemitsu S, Nakashima H, Otsuka T. Association of CTLA-4 but not CD28 gene polymorphisms with systemic lupus erythematosus in the Japanese population. Rheumatology (Oxford). 2001;40(6):662–667
- Matsushita M, Tsuchiya N, Shiota M, et al. Lack of a strong association of CTLA-4 exon 1 polymorphism with the susceptibility to rheumatoid arthritis and systemic lupus erythematosus in Japanese: An association study using a novel variation screening method. Tissue Antigens. 1999;54(6):578–584
- Takeuchi F, Kawasugi K, Nabeta H, Mori M, Tanimoto K. CTLA-4 dimorphisms in Japanese patients with systemic lupus erythematosus. Clin Exp Rheumatol. 2003;21(4):527–528
- Aguilar F, Torres B, Sánchez-Román J, Núñez-Roldán A, González-Escribano MF. CTLA-4 polymorphism in Spanish patients with systemic lupus erythematosus. Hum Immunol. 2003;64(10):936–940
- Barreto M, Santos E, Ferreira R, et al. Evidence for CTLA4 as a susceptibility gene for systemic lupus erythematosus. Eur J Hum Genet. 2004;12(8):620–626
- Heward J, Gordon C, Allahabadia A, Barnett AH, Franklyn JA, Gough SC. The A-G polymorphism in exon 1 of the CTLA-4 gene is not associated with systemic lupus erythematosus. Ann Rheum Dis. 1999;58(3):193–195
- Brozzetti A, Marzotti S, Tortoioli C, et al. Cytotoxic T lymphocyte antigen-4 Ala17 polymorphism is a genetic marker of autoimmune adrenal insufficiency: Italian association study and meta-analysis of European studies. Eur J Endocrinol. 2010;162(2):361–369
- Ligers A, Teleshova N, Masterman T, Huang W, Hillert J. CTLA-4 gene expression is influenced by promoter and exon 1 polymorphisms. Genes Immun. 2001;2(3):145–152
- Sun T, Zhou Y, Yang M, et al. Functional genetic variations in cytotoxic T-lymphocyte antigen 4 and susceptibility to multiple types of cancer. Cancer Res. 2008;68(3):7025–7034
- Nistico L, Buzzetti R, Pritchard LE, et al. The CTLA-4 gene region of chromosome 2q33 is linked to, and associated with, type 1 diabetes. Hum Mol Genet. 1996;5:1075–1080
- Kavvoura FK, Akamizu T, Awata T, et al. Cytotoxic T-lymphocyte associatedantigen 4 gene polymorphisms and autoimmune thyroid disease: A meta-analysis. J Clin Endocrinol Metab. 2007;92(7):3162–3170
- Munoz-Valle JF, Valle Y, Padilla-Gutierrez JR, et al. The +49A>G CTLA-4 polymorphism is associated with rheumatoid arthritis in Mexican population. Clin Chim Acta. 2010;411(9–10):725–728
- Miyake Y, Ikeda F, Takaki A, Nouso K, Yamamoto K. +49A/G polymorphism of cytotoxic T-lymphocyte antigen 4 gene in type 1 autoimmune hepatitis and primary biliary cirrhosis: A meta-analysis. Hepatol Res. 2011;41(2):151–159
- Thornton A, Lee P. Publication bias in meta-analysis: Its causes and consequences. J Clin Epidemiol. 2000;53(2):207–221