Abstract
Background: Genetic polymorphism of endothelial nitric oxide synthase (eNOS) has been implicated in the risk of diabetic nephropathy (DN), but the published findings were inconsistent. We performed a comprehensive meta-analysis to derive a more precise estimation of the association between genetic polymorphisms of eNOS and the risk of DN. Methods: Six online database were researched on the associations between polymorphisms of eNOS (T786C, G894T, 4b/4a) and DN risk. PRISMA statement and Hardy–Weinberg equilibrium assessment were used in this report. Odds ratio and 95% confidence interval were estimated based on the following genetic contrast/models: allelic model, dominant model, recessive model and co-dominant model. The publication bias and sensitivity analysis were also performed to guarantee the statistical power. Results: A total of 49 case–control studies with 11,990/9754/5131 participants for DN/DM/HC group were eligible for meta-analysis (7/25/31 studies for T786C/G984T/4b/a). For the eNOS-T786C, C allele showed a weak association between C allele and DN risk in DN/T2DM group. For eNOS-G894T, there was an association between T allele and DN risk in the global, Asian and African population in DN/T2DM group. For the eNOS-4b/4a, 4a allele was found contributing significantly to increased DN risk in the global population. Conclusion: Our comprehensive meta-analysis suggests that three polymorphisms of eNOS may be the increased risk factors of DN development, especially in Asian population and T2DM group.
Introduction
Diabetic nephropathy (DN) is a common microvascular complication of diabetes.Citation1 DN is the leading cause of end-stage renal disease worldwide and is the second leading cause of blood dialysis in China (13.5%). The prevalence of diabetes is increasing globally, with India, China and the USA being the top three countries estimated to have the highest number of people with diabetes. The reason why some diabetics develop nephropathy while others do not despite having long-term hyperglycemia remains unknown. Thus, researchers have explored the genetic background of the host to explain this phenomenon.
Nitric oxide (NO) is a ubiquitous vasodilator and an important regulator of renal sodium excretion.Citation2 NO is produced by the oxidation of l-arginine to l-citrulline by NO synthase (NOS). NOS has three isoforms: neuronal NOS, inducible NOS and (constitutive) endothelial NOS (eNOS or NOS3).Citation3,Citation4 Decreased NO activity can be attributed to the impaired production of NO due to uncoupled receptor-mediated signal transduction, deficient NOS substrate l-arginine, or decreased availability of one or more cofactors essential for the optimal function of NOS.Citation5 Reduced NO generation induces renal injury,Citation6 and impaired endothelial NO generation is the major deterioration factor for progressive renal disease.
Current investigations suggested that gene polymorphisms play a key role in the onset of DN. Many genes have been associated with DN. For instance, eNOS regulates vascular function during DN. The eNOS gene is located on chromosome 7q35–36, comprises 26 exons and 25 introns that span 21 kb, and is primarily expressed in the endothelium.Citation7 The eNOS gene encodes a 1.5 kDa protein composed of 1204 amino acids. Previous studies have investigated three eNOS polymorphisms, namely, the T786C single-nucleotide polymorphism, the G894T (Glu298Asp) missense mutation in exon 7 and the intron 427-bp repeat (4b/a) in the promoter region. T786C reduces eNOS transcription, G894T has been associated with decreased eNOS activity and 4b/a has been associated with decreased plasma NO concentrations that may reflect eNOS activity.
Some reports have confirmed the relationship between eNOS genetic polymorphisms and DN. However, the pathogenesis of the development and progression of DN remains unclear to date. In addition, the biological, pathological and functional mechanisms responsible for the potential association of eNOS polymorphisms with DN risk have yet to be elucidated in detail. Therefore, the present meta-analysis examined the eNOS T786C, G894T, and 4b/a polymorphisms and their relationship to DN risk.
Methods
Search strategy
We conducted a computerized literature search up to October 2014 for case–control studies to identify all published studies that examined the association of eNOS genetic polymorphisms (T786C, G894T and 4b/4a) with DN risk. The databases used in the present study were as follows: PubMed, Web of Science, Embase, China Knowledge Resource Integrated, VIP, Wanfang and China Biological Medicine.
The following keywords were used: (“NOS” or “eNOS” or “NOS3” or “ecNOS” or “endothelial nitric oxide synthase” or “nitric oxide synthase type III” or “T786C” or “Glu298Asp” or “G894T” or “4b/a”) and (“gene” or “gene polymorphism” or “genetic polymorphism” or “polymorphisms” or “variant”) and (“diabetic” or “diabetes mellitus” or “diabetic nephropathy” or “diabetic kidney disease” or “diabetes mellitus nephropathy” or “kidney in diabetes” or “chronic renal insufficiency”). In the CNKI, VIP and Wanfang databases, the corresponding Chinese characters of the keywords were used in the search. We also extended the search spectrum to the “related articles” and the bibliographies of all retrieved studies. The retrieved abstracts were read to identify studies that examined the allele frequencies and genotype associations between eNOS polymorphisms and DN. In addition, all references cited were reviewed to identify additional studies. Only English and Chinese language publications were acknowledged.
Inclusion criteria
Studies were included in this meta-analysis according to the following criteria: (1) case–control studies on the association between eNOS polymorphisms and DN risk; (2) data were available on genotype distributions for calculating the odds ratio (OR) with its 95% confidence interval (CI), or sufficient data were provided to calculate the corresponding estimate effect [OR, (95% CI)]; (3) at least two comparison groups [DN group and/or diabetes mellitus (DM) group and/or healthy controls] were used. When multiple publications of the same data from the same study group occurred, we only included the most recent or complete paper for analysis. When a study reported results on different subgroups, we treated each subgroup as a single comparison in the meta-analysis.
Exclusion criteria
Studies were excluded from the meta-analysis according to the following criteria: (1) review articles, editorials and case reports; (2) articles that did not provide detailed genotype data; (3) articles that investigated the association of other genes with DN and (4) articles that investigated the role of eNOS in other diseases.
Data extraction and synthesis
Two investigators independently conducted the search, extracted and tabulated all the relevant data, and reached a consensus on all the items. The following information was independently extracted from the identified studies: surname of the first author, year of publication, ethnicity of the study population, language, type of DM, sample size, gender, age, DM duration, body mass index, HbA1C level, genotype distribution and allele frequencies in case/control groups for the eNOS polymorphisms. Subjects were classified into three groups: DN group (DM with DN), DM group (DM without DN) and HC group (healthy individuals including volunteers, hospital staff, etc.). Frequencies of alleles were calculated for case/control groups from the corresponding genotype distributions. Discrepancies were resolved by discussion and consensus with all the authors. We also tested the Hardy–Weinberg equilibrium (HWE) of genotype distributions in the DM/HC group.
Statistical analysis
We examined eNOS genotypes under the allelic, dominant, recessive, additive and codominant models. The meta-analysis was conducted using the Review Manager 5.1 software (Cochrane IKMD, London, UK). The total ORs with 95% CIs of the forest plot were estimated to evaluate the strength of the association between eNOS polymorphisms and DN risk. Statistical significance was considered at p < 0.05 for the overall ORs. A funnel plot was used to evaluate the publication bias in our meta-analysis.
Heterogeneity assumption was assessed by chi-square-based Q test and I2 test.Citation8,Citation9 The heterogeneity was considered statistically significant when p < 0.10. I2 < 50% denoted a non-significant heterogeneity among the involved studies in the meta-analysis. The fixed-effects model (Mantel–Haenszel method, if p ≥ 0.10) or the random-effects model (DerSimonian and Laird method, if p < 0.10) was applied to summarize the combined ORs.Citation10 These two models provide similar results when heterogeneity between studies was absent; otherwise, the random-effects model is appropriate. Sensitivity analysis was performed when studies with DM and HC groups were not in HWE.
Results
Subjects
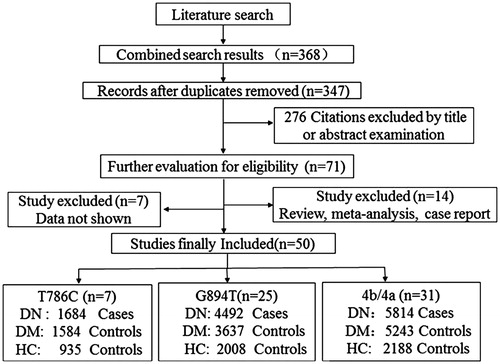
A flowchart of the literature search and study selection is illustrated in . A total of 368 articles were identified on the basis of our search criterion. After removing duplicated publications, we harvested 347 articles, among which 276 were filtered by title or abstract. Among the remaining 71 studies, 7 were excluded because of insufficient information about genotype distributions and 14 were excluded because of being review articles, meta-analyses and case reports. The subjects in the present meta-analysis were classified into three groups: DN group (DN patients), DM group (DM patients without DN) and HC group (healthy individuals including volunteers, hospital staff, and so on).
Finally, 50 case–control studiesCitation11–60 with three polymorphisms fulfilled the eligible criteria and were included in the current meta-analysis (). This meta-analysis involved 7 studies on T786C (1684, 1584 and 935 DN, DM and HC subjects, respectively), 25 studies on G894T (4492, 3637 and 2008 DN, DM and HC subjects, respectively) and 31 studies on 4b/4a (5814, 5243 and 2188 DN, DM and HC subjects, respectively). The detailed characteristics of each study are listed in . These studies were conducted in different populations of various ethnicities: 35 studies on Asians, 12 studies on Caucasians and 3 studies on Africans. A total of 32 studies were reported in English, whereas 19 reports were published in Chinese. Eleven studies with controls (one study for T-786 C, four studies for G984T and six studies for 4b/a) were not in HWE (); therefore, these studies were subjected to a sensitivity analysis.
Table 1. Characteristics of studies included in the meta-analysis.
Table 2. Distribution of eNOS polymorphisms and frequency of allele for studies.
Association of the eNOS-T786C polymorphism with DN risk
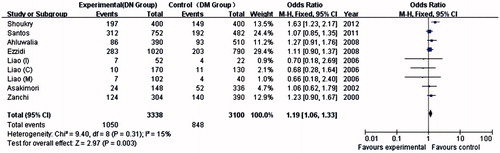
A total of 4203 subjects, which included 1684 DN patients, 1584 DM patients and 935 healthy controls in seven studies, were investigated for eNOS-T786C (). Three, two and two of the seven eligible studies focused on Asians, Caucasians and Africans, respectively. Among these studies, one concentrated on type 1 DM (T1DM), five on type 2 (T2DM), and one was not applicable.
In the allelic analysis, no significant heterogeneity was found among the studies (DN/DM as cases/controls; PQ = 0.31, I2 = 15%). In the genotype analysis, heterogeneity existed in both the recessive model (PQ = 0.01, I2 = 66%) and the codominant (CC vs. TT) model (PQ = 0.02, I2 = 64%) for the association of the T786C polymorphism.
Only seven studies investigated the association between eNOS-T786C polymorphisms and DN risk. As shown in , the association between the C allele and DN risk was weaker than that between the T allele and DN risk in the global population (p = 0.003, OR = 1.19, 95% CI = 1.06–1.33). No association was detected between the CC genotype and DN risk in the global population in the recessive genetic model (p = 0.1, OR = 1.48, 95% CI = 0.93–2.38) or in the codominant model (CC vs. TT, TC vs. TT). Meanwhile, only a weak association was found between the CC genotype and DN risk in the dominant model (p = 0.03, OR = 1.18, 95% CI = 1.02–1.37).
Heterogeneity existed in all models (DN/HC group as cases/controls), and no association was found with DN among these studies (). In the sensitivity analysis, no study produced a p-value above 0.05, and no study was excluded in this meta-analysis.
Table 3. Meta-analysis for eNOS-T786C Polymorphism.
Heterogeneity existed in all models (DM/HC group as cases/controls), except for the recessive model (PQ = 0.18, I2 = 45%), and the allele frequencies and genotype in the DM patients were not significantly different from those in the healthy individuals in all genetic models, except for the recessive model (p = 0.007, OR = 1.93, 95% CI = 1.20–3.10). Furthermore, no study was excluded after the sensitivity analysis in this subgroup analysis.
Association of the eNOS-G894T polymorphism with DN risk
A total of 25 studies that included 10,137 subjects (4492 DN patients, 3637 DM patients and 2008 healthy controls) were used in our analysis (). Among the 25 eligible studies, 18 focused on Asians, 4 on Caucasians and 3 on Africans. We performed a subgroup analysis according to the cases/controls by the DN/DM group (ethnicity and type of DM), DN/HC group, DM/HC group and DN clinical stages (microalbuminuria and macroalbuminuria).
For the ethnicity subgroup of eNOS-G894T (DN/DM as cases/controls), 15 studies were conducted on Asians, 4 on Caucasians, 3 on Africans and 3 were not applicable. A significant heterogeneity was detected in the allelic model for the global population (PQ = 0.00001, I2 = 72%), Asian population (PQ = 0.0007, I2 = 61%) and African population (PQ = 0.1, I2 =57%) but not for the Caucasian population (PQ = 0.81, I2 = 0%). An association was found between the T allele and DN risk compared with the G allele in the global population (p = 0.0001, OR = 1.37, 95% CI = 1.17–1.61), Asian population (p < 0.0001, OR = 1.64, 95% CI = 1.29–2.08), and African population (p = 0.004, OR = 1.52, 95% CI = 1.15–2.03) but not in the Caucasian population (p = 0.17, OR = 0.92, 95% CI = 0.83–1.03). However, in the ethnicity of genotype analysis, no significant heterogeneity existed in the Caucasians and Africans in all models (). Meanwhile, no association was found between the eNOS-G894T TT genotype and DN risk in the Caucasians in all models. By contrast, a significant result was observed in the African population in all models ().
Figure 3. Funnel plots for the allelic model by ethnicity in DN/DM group: A, B and C for T786C, G894T and 4b/4a, respectively.
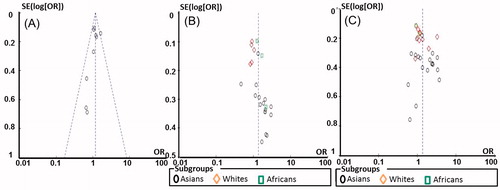
Table 4. Meta-analysis for eNOS-G894T Polymorphism.
For the DM subgroup of eNOS-G894T (DN/DM as cases/controls), 2 and 20 studies concentrated on T1DM and T2DM, respectively, and 3 studies were not applicable (). In the meta-analysis, no association was detected between the eNOS-G894T polymorphisms (T allele frequencies and TT genotype) and DN risk in T1DM in all models. By contrast, a significant association was observed in T2DM in all models, except for the codominant model (TT vs. GG; p = 0.12, OR = 1.38, 95% CI = 0.92–2.07).
For the subgroup of clinical stages (microalbuminuria and macroalbuminuria) with four eligible studies, no association was found between eNOS-G894T (T allele frequencies and TT genotype) and DN risk in all models. By contrast, the T allele frequencies and TT genotype were higher in the DN group than in the HC group in all genetic models in the DN/HC subgroup analysis (). Meanwhile, no significant heterogeneity existed in the recessive and codominant genetic models (TT vs. GG) between the two subgroups under the fixed-effects model.
For the subgroup of DM/HC subjects (cases/controls) with 13 eligible studies, no significant heterogeneity existed in all genetic models under the fixed-effects model. Furthermore, no association was found between eNOS-G894T (T allele frequencies and TT genotype) and DN risk in all genetic models.
Association of the eNOS-4b/4a polymorphism with DN risk
A total of 13,245 subjects (5814 DN patients, 5243 DM patients and 2188 healthy controls) were involved in the eNOS-4b/4a polymorphisms with 31 studies (). In all eligible studies, we performed subgroup analyses according to the cases/controls by the DN/DM group (ethnicity and type of DM), DN/HC group, DM/HC group and DN clinical stages (microalbuminuria and macroalbuminuria, macroalbuminuria and advanced nephropathy).
For the ethnicity subgroup of eNOS-4b/4a (DN/DM as cases/controls; ), 18 studies were conducted on Asians, 8 on Caucasians, 2 on Africans and 3 were not applicable. A significant heterogeneity existed among the studies, except in the African population (PQ = 0.26, I2 = 22%), in the allelic model analysis. The 4a allele of eNOS-4b/4a significantly contributed to increased DN risk in the global population (p < 0.0001, OR = 1.35, 95% CI = 1.16–1.57), Asian population (p = 0.0006, OR = 1.50, 95% CI = 1.19–1.89) and Caucasian population (p = 0.04, OR = 1.28, 95% CI = 1.01–1.62) but not in the African population (p = 0.98, OR = 1.00, 95% CI = 0.80–1.24). Significant associations existed between 4a/4a genotype distributions and DN risk in the global and Asian populations in all models. However, no association was found in the African population in all models ( and ).
Table 5. Meta-analysis for eNOS-4b/4a polymorphism.
Table 6. Scale for quality assessment.
Table 7. Search History by example of the PubMed.
For the DM type subgroup of eNOS-4b/4a (DN/DM as cases/controls), 6 and 22 studies concentrated on T1DM and T2DM, respectively, and 3 studies were not applicable. In the subgroup analysis, no significant associations existed between eNOS-4b/4a polymorphisms (4a allele frequencies and 4a/4a genotype distributions) and DN risk in T1DM in all models. By contrast, a significant association was observed in T2DM in all models, except for the codominant model (4b/4a vs. 4b/4b; p = 0.06, OR = 1.21, 95% CI = 0.99–1.49).
For the subgroup of DN clinical stages (microalbuminuria vs. macroalbuminuria with seven studies, macroalbuminuria vs. advanced nephropathy with three studies), no associations existed between eNOS-4b/4a polymorphisms (4a allele frequencies and 4a/4a genotype distributions) and DN risk in all models at any DN clinical stage without heterogeneity among the eligible studies ().
For the DN/HC subgroup (cases/controls) with 15 studies, no heterogeneities were found in the recessive and codominant genetic models under the fixed-effects model, except in the allelic and dominant genetic models. As shown in , the 4a allele frequencies and 4a/4a genotype distributions were significantly higher in the DN group than in the HC group in all models ().
For the DM/HC subgroup (cases/controls) with 12 eligible studies, heterogeneity existed in the allelic and dominant genetic models under the random-effects model. Thus, a sensitivity analysis was performed. The meta-analysis revealed that 4a allele frequencies were significantly higher in the DM group than in the HC group (p = 0.03, OR = 1.36, 95% CI = 1.02–1.80). Meanwhile, the 4a/4a genotype distributions of eNOS-4b/4a displayed significant associations in all genetic models, except for the dominant model (p = 0.08, OR = 1.30, 95% CI = 0.97–1.74; ).
Discussion
This meta-analysis involving 11,990 DN patients, 10,464 DM controls and 5131 healthy controls from 50 independent publications examined the associations of three eNOS gene polymorphisms (T786C, G894T and 4b/4a) with DN risk. Several scholars have performed meta-analyses in recent years, and their variable study designs, ethnicities and control sources produced controversial conclusions. Therefore, we performed an updated meta-analysis to explore the pathological, biological and functional mechanisms of the eNOS polymorphisms in DN.
For eNOS-T786C polymorphisms, the C allele was associated with an increased risk of DN in our meta-analysis, whereas the CC genotype failed to demonstrate any obvious association. In relation to the eNOS-T786C polymorphism in the promoter region, the C allele has been associated with an increased risk of DN in North Asian IndiansCitation11 and with end-stage renal disease (ESRD) in Japanese.Citation17 However, some studies did not observe an association between this polymorphism and renal insufficiency in TunisiansCitation14 and in North and South Asian IndiansCitation21 with T2DM. The T-786 C variant has been suspected to reduce the promoter activity in eNOS by 50%, thereby reducing eNOS mRNA accumulation and inducing NO production. However, we only included seven studies in our meta-analysis. Therefore, the results should be interpreted with caution. The sensitivity analysis for HWE in T786C did not alter the pattern of results.
No association was found with DN in the DN/HC group. We only extracted two characteristics from the eligible studies in this meta-analysis. The small sample size and the varied geographic/ethnic distributions causing the heterogeneity contributed to the finding. Therefore, further studies with large sample sizes for quantitative traits in eNOS-T786C must be conducted to achieve conclusive conclusions.
For eNOS-G894T polymorphisms, our meta-analysis manifested that the T allele and TT genotype were associated with the onset of DN in the African population, and the T allele was positively associated with DN in the Asian population. However, the T allele and TT genotype showed no association with DN in the Caucasian population. Furthermore, the GG genotype seemed to play a protective role against DN risk. A sensitivity analysis was also performed in our meta-analysis. The results of the sensitivity analysis were similar to those of the non-sensitivity analysis for overall populations. The results for the overall populations might be robust to some extent. The geographic and ethnic differences might affect the association of the gene polymorphisms with DN risk. One explanation is that G894T may not have a significant effect on the disease, and the allele associated with the real causative mutations varies across ethnic groups. Another possible explanation is that the variant may influence the disease, but the genetic backgrounds of the different populations (e.g., different properties of other proteins in related pathways) may result in different responses and outcomes. Distinct living environments, habits and diets may also mediate the results.
However, the T allele and TT genotype showed no association in the Caucasian population. These findings should be interpreted with caution because factors such as heterogeneity of enrolled cases, limited statistical power, variable study designs, different ethnicities (Asians, Caucasians and Africans), DN clinical stages (microalbuminuria, macroalbuminuria and ESRD), DM type (type 1 and type 2), control source (DM group and healthy controls) and different interventions might exert substantial effects on between-study heterogeneity.
The present study analyzed the distribution of genetic variants of the eNOS intron 4 polymorphism in DN and DM patients. The frequency of the eNOS 4a allele carriers was significantly higher in the DN group than in the T2DM group, indicating that the eNOS 4a allele may be a risk factor for DN in the Asian and Caucasian populations but not in the African population. The present results were supported by the findings of Chinese,Citation39,Citation42–44 Japanese,Citation17,Citation52 and Iranian scholars.Citation58 However, other studies concluded that the 4a allele is not associated with DN. Lin et al. Citation47 concluded that the a-allele in intron 4 of the eNOS gene does not increase the risk of DN in Chinese T2DM patients. Fujita et al.Citation51 and Shimizu et al.Citation45 also found no association between the 4a allele gene and DN in East Asian and Japanese patients, respectively. Moreover, ShoukryCitation11 showed no significant difference in 4a allele and DN in Egyptian subjects.
In a recent meta-analysis by He et al., eNOS-4b/4a polymorphisms (4a vs. 4b allele) were significantly associated with developing DN in Asians but not in Caucasians. The specific mechanism has not yet been clarified, and the available data have been controversial. Tsukada et al.Citation61 demonstrated that the 4a allele is significantly associated with decreasing NO metabolite levels in plasma. Therefore, the important mechanisms underlying the occurrence of DN might be interpreted by the b-insertion/a-deletion polymorphism in intron 4 of the eNOS gene. In particular, this polymorphism could downregulate eNOS expression and thus decreases NO production. Hence, we cannot exclude the possibility of the 4b/a variable number of tandem repeats as a functional significant factor to DN risk.
The present results indicated that eNOS 4a/b had a higher distribution in the DN patients than in the T2DM patients in the global and Asian populations but not in the African population. The results were consistent with previously reported findings on different populations.Citation53 However, some studies did not observe any association.Citation42 This phenomenon may be ascribed to different reasons. One explanation for the discrepancy may be the ethnicity of the study populations. The incidence of DN sharply increased in Asians as compared with other populations, which suggested a distinguishable background of pathology in the Asian population. Such a background may be genetic such as different linkage maps or due to different lifestyles or public sanitation. Further ethnic-specific association studies are warranted to elucidate the population discrepancy of DN risk factors. Another explanation for the discrepancy may be genetic heterogeneity. The current data revealed that the frequency of each genotype significantly differed among the populations and that this heterogeneity may statistically contribute to the discrepancy of pooled OR.
In the present meta-analysis, a significant association was observed between eNOS polymorphisms and DN. This association was enhanced when subgroup studies were performed among T2DM patients. Basing on these results, we conclude that eNOS (4a allele and 4a/4a genotype) is an effective genetic factor that contributes to the pathology of DN. This conclusion is consistent with published functional research on the eNOS gene.Citation59
In the present study, eNOS-4b/4a (4a allele and 4a/4a) or eNOS-G894T (T allele and TT genotype) did not show an association with DN risk at any clinical stage in all models. Therefore, we assumed that this polymorphism could be an aggravating factor rather than an initiating factor for DN.
The findings of our meta-analysis should be interpreted with caution because of the following limitations. (1) Although we set a sensitive search strategy for the retrieval of eligible studies, some studies might have been inevitably lost. (2) This meta-analysis only involved Chinese and English published studies. Therefore, non-English, non-Chinese, and non-published studies were not reviewed, which might induce language bias. (3) Several papers were excluded from our study because we could not obtain the original genotype numbers by email from the authors. Therefore, we lost the opportunity to obtain a larger sample size and increased statistical power.
In conclusion, the allele frequencies and genotype distributions of eNOS-T786C, eNOS-G894T and eNOS-4a/b polymorphisms significantly contributed to DN risk, but the eNOS-T786C CC genotype distribution was not associated with DN in global populations. The eNOS-G894T and eNOS-4a/b polymorphisms had higher risk in T2DM patients than in T1DM patients. The eNOS-G894T and eNOS-4b/4a polymorphisms did not show an association with DN risk at any clinical stage. These conclusions highlighted the importance of the three polymorphisms on DN, which might be a useful suggestion for clinical therapeutic intervention.
Acknowledgments
The authors would like to thank Drs. Xian-Tao Zeng and Hong-Xia Zuo (Center for Evidence-Based Medicine and Clinical Research, Taihe Hospital, Hubei University of Medicine) for the invaluable discussions about the study design and statistical analyses.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
References
- Lee AS, Lee YJ, Lee SM, et al. An aqueous extract of Portulaca oleracea ameliorates diabetic nephropathy through suppression of renal fibrosis and inflammation in diabetic db/db mice. Am J Chin Med. 2012;40(3):495–510
- Larsen T, Mose FH, Bech JN, Pedersen EB. Effect of nitric oxide inhibition on blood pressure and renal sodium handling: A dose–response study in healthy man. Clin Exp Hypertens. 2012;4(8):567–574
- Moncada S, Higgs EA. The discovery of nitric oxide and its role in vascular biology. Br J Pharmacol. 2006;147(Suppl 1):S193–S201
- Wang Y, Marsden PA. Nitric oxide synthases: Biochemical and molecular regulation. Curr Opin Nephrol Hypertens. 1995;4(1):12–22
- Honing ML, Morrison PJ, Banga JD, Stroes ES, Rabelink TJ. Nitric oxide availability in diabetes mellitus. Diabetes Metab Rev. 1998;14(3):241–249
- Schwartz IF, Grupper A, Soetendorp H, et al. Attenuated glomerular arginine transport prevents hyperfiltration and induces HIF-1α in the pregnant uremic rat. Am J Physiol Renal Physiol. 2012;303(3):F396–F404
- Marsden PA, Heng HH, Scherer SW, et al. Structure and chromosomal localization of the human constitutive endothelial nitric oxide synthase gene. J Biol Chem. 1993;268(23):17478–17488
- Coory MD. Comment on: Heterogeneity in meta-analysis should be expected and appropriately quantified. Int J Epidemiol. 2010; 39(3):932–933
- Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558
- DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188
- Shoukry A, Shalaby SM, Abdelazim S, et al. Endothelial nitric oxide synthase gene polymorphisms and the risk of diabetic nephropathy in type 2 diabetes mellitus. Genet Test Mol Biomarkers. 2012;16(6):574–579
- Santos KG, Crispim D, Canani LH, Ferrugem PT, Gross JL, Roisenberg I. Association of eNOS gene polymorphisms with renal disease in Caucasians with type 2 diabetes. Diabetes Res Clin Pract. 2011;91(3):353–362
- Ahluwalia TS, Ahuja M, Rai TS, et al. Endothelial nitric oxide synthase gene haplotypes and diabetic nephropathy among Asian Indians. Mol Cell Biochem. 2008;314(1–2):9–17
- Ezzidi I, Mtiraoui N, Mohamed MB, Mahjoub T, Kacem M, Almawi WY. Association of endothelial nitric oxide synthase Glu298Asp, 4b/a, and -786T>C gene variants with diabetic nephropathy. J Diabetes Complications. 2008;22(5):331–338
- Zanchi A, Moczulski DK, Hanna LS, Wantman M, Warram JH, Krolewski AS. Risk of advanced diabetic nephropathy in type 1 diabetes is associated with endothelial nitric oxide synthase gene polymorphism. Kidney Int. 2000;57(2):405–413
- Liao L, Zhao JJ, Lin MJ, Chen SW, Li GA. Association of a polymorphism in endothelial nitric oxide synthase gene T-786→C with diabetic nephropathy. Chin J Lab Med. 2006;29(2):146–147
- Asakimori Y, Yorioka N, Yamamoto I, et al. Endothelial nitric oxide synthase intron 4 polymorphism influences the progression of renal disease. Nephron. 2001;89(2):219–223
- Rahimi Z, Vaisi-Raygani A, Rahimi Z, Parsian A. Concomitant presence of endothelial nitric oxide 894T and angiotensin II-converting enzyme D alleles are associated with diabetic nephropathy in a Kurdish population from Western Iran. Nephrology (Carlton). 2012;17(2):175–181
- Rao GF, Chen EF, Lin HY, Wu JY, Zeng AP. The correlation of eNOS gene polymorphism and susceptibility to diabetic nephropathy. China Modern Doctor. 2012;50(7):49–50
- Dai HS, Zhang Y. An association study of MTHFR and eNOS genes polymorphism with diabetic nephropathy. Chin J Trauma Disabil Med. 2012;20(6):4–6
- Tiwari AK, Prasad P, Thelma BK, et al. Oxidative stress pathway genes and chronic renal insufficiency in Asian Indians with Type 2 diabetes. J Diabetes Complications. 2009;23(2):102–111
- Tamemoto H, Ishikawa SE, Kawakami M. Association of the Glu298Asp polymorphism of the eNOS Gene with ischemic heart disease in Japanese diabetic subjects. Diabetes Res Clin Pract. 2008;80(2):275–279
- Ma JR, Yu DM, Liu DM. Study on the relationship between G894T mutation in endothelial constitutive nitric oxide synthase (ecNOS) gene and diabetic nephropathy. Tianjin Med J. 2006; 34(8):527–530
- Dong YH, Qu SP, Lv WS, et al. Gene polymorphism in chromosome 7q35 and susceptibility to diabetic nephropathy. Chin J Endocrinol Metab. 2005;21(1):47–50
- Fu ZJ, Li CG, Wang ZC, Yan SL. Coexistence of aldose reduction gene and endothelial nitric oxide synthase polymorphisms associates with diabetic nephropathy. J Clin Rehabil Tissue Eng Res. 2007;11(34):6893–6896
- Shin Shin Y, Baek SH, Chang KY, et al. Relations between eNOS Glu298Asp polymorphism and progression of diabetic nephropathy. Diabetes Res Clin Pract. 2004;65(3):257–265
- Li CG, Lv SY, Qi YQ, Wang L, Yan SL. Association study of 894G→T mutation at exon 7 of endothelial nitric oxide synthase gene and diabetic nephropathy with hypertension. Acta Acad Med Qingdao Univ. 2004;40(3):200–202
- Lv SY, Xing QQ, Li CG. Association between 894G-T polymorphism at exon 7 of endothelial nitric oxide synthase gene and diabetic nephropathy with or without hypertention. Chin J Mult Organ Dis Elderly. 2003;2(4):275–311
- Lv WS, Dong YH, Wang HY, Liu L, Si YG. A study of the association between polymorphism of endothelial nitric oxide synthase gene and diabetic nephropathy. Chin J Diabetes. 2002;10(2):81–84
- Möllsten A, Wessman M, Svensson M, et al. Glu298Asp and NOS4ab polymorphisms in diabetic nephropathy. Ann Med. 2006;38(7):522–528
- Möllsten A, Lajer M, Jorsal A, Tarnow L. The endothelial nitric oxide synthase gene and risk of diabetic nephropathy and development of cardiovascular disease in type 1 diabetes. Mol Genet Metab. 2009;97(1):80–84
- Cai H, Wang X, Colagiuri S, Wilcken DE. A common Glu298–>Asp (894G–>T) mutation at exon 7 of the endothelial nitric oxide synthase gene and vascular complications in type 2 diabetes. Diabetes Care. 1998;21(12):2195–2196
- El-Din Bessa SS, Hamdy SM. Impact of nitric oxide synthase Glu298Asp polymorphism on the development of end-stage renal disease in type 2 diabetic Egyptian patients. Ren Fail. 2011;33(9):878–884
- Li XN, Yu MX, Wu XY, Shui H, Xiao JS. Association of endothelial nitric oxide synthase gene Glu298Asp polymorphism with diabetic kidney disease. J Clin Nephrol. 2011;11(8):351–353
- Wang ZC. The Study of the Relationship Between Glu298Asp (894G→T) Single Nucleotide Polymorphisims at Exon 7 of the Human Endothelial Cell Nitric Oxide Synthase (heNOS) Gene and Diabetic Nephropathy. Qingdao, China:QingDao University; 2004
- Nagase S, Suzuki H, Wang Y, et al. Association of ecNOS gene polymorphisms with end stage renal diseases. Mol Cell Biochem. 2003;244(1–2):113–118
- Thaha M, Pranawa, Yogiantoro M, Sutjipto, et al. Association of endothelial nitric oxide synthase Glu298Asp polymorphism with end-stage renal disease. Clin Nephrol. 2008;70(2):144–154
- Suzuki H, Nagase S, Kikuchi S, Wang Y, Koyama A. Association of a missense Glu298Asp mutation of the endothelial nitric oxide synthase gene with end stage renal disease. Clin Chem. 2000;46(11):1858–1860
- Guo XJ, Liu SJ. The association between endothelial nitric oxide synthase gene polymorphism and type 2 diabetic nephropathy. Natl Med Front China. 2011;6(19):3–5
- Dong JJ, Zhao JJ, Lee KO, Lim MC, Chan SW, Liao L. Distribution of polymorphism in endothelial nitric oxide synthase gene in mainland Chinese and Singapore Chinese and its association with diabetic nephropathy. Natl Med J China. 2007;87(48):3415–3417
- Zhang M, Liu H, Yang HY, Wang YM, Song DP, Li H. Relationship of endothelial nitric oxide synthase (eNOS) 4a/b gene polymorphism with diabetic nephropathy in T2DM. Chin J Diabetes. 2005;13(4):284–285
- Xing Q, Su BL, Li CC, et al. Association between endothelial nitric oxide synthase gene polymorphism and type 2 diabetic nephropathy. Chin J Endocrinol Metab. 2004;20(5):435–437
- Sun HY, Yang MG, Liu SQ, et al. Study on the correlation of the polymorphisms of endothelial nitric oxide synthase gene with type 2 diabetes mellitus and diabetic nephropathy. Chin J Prev Contr Chron Non-commun Dis. 2004;12(3):101–103
- Luo H, Ning YY. Relationship between gene polymorphism of endothelial nitricoxide synthase and early intervention of patients with diabetic nephropathy. Chin J Clin Rehabil. 2003;7(12):1766–1767
- Shimizu T, Onuma T, Kawamori R, Makita Y, Tomino Y. Endothelial nitric oxide synthase gene and the development of diabetic nephropathy. Diabetes Res Clin Pract. 2002;58(3):179–185
- Li CG, Yang NL. The correlation between PAI-1 promoter 4G/5G and the intron 4 of eNOS gene polymorphism and diabetic nephropathy. Shandong Med J. 2002;42(17):16–18
- Lin S, Qu H, Qiu M. Allele A in intron 4 of ecNOS gene will not increase the risk of diabetic nephropathy in type 2 diabetes of Chinese population. Nephron. 2002;91(4):768
- Taniwaki H, Ishimura E, Matsumoto N, Emoto M, Inaba M, Nishizawa Y. Relations between ACE gene and ecNOS gene polymorphisms and resistive index in type 2 diabetic patients with nephropathy. Diabetes Care 2001;24(9):1653–1660
- Lv WS, Dong YH, Li CG, Liu L, Wang HY, Si YG. A study of association between polymorphism of endothelial nitric oxide synthase gene and diabetic nephropathy. J Weifang Med Coll. 2001;23(4):278–280
- Asakimori Y, Yorioka N, Taniguchi Y, et al. T(-786)–>C polymorphism of the endothelial nitric oxide synthase gene influences the progression of renal disease. Nephron. 2002;91(4):747–751
- Fujita H, Narita T, Meguro H, et al. Lack of association between an ecNOS gene polymorphism and diabetic nephropathy in type 2 diabetic patients with proliferative diabetic retinopathy. Horm Metab Res. 2000;32(2):80–83
- Neugebauer S, Baba T, Watanabe T. Association of the nitric oxide synthase gene polymorphism with an increased risk for progression to diabetic nephropathy in type 2 diabetes. Diabetes. 2000;49(3):500–503
- Shestakova MV, Koshel' LV, Vagodin VA, Dedov II. Risk factors of diabetic nephropathy progression in patients with a long history of diabetic mellitus as shown by a retrospective analysis. Ter Arkh. 2006;78(5):60–64
- Rippin JD, Patel A, Belyaev ND, Gill GV, Barnett AH, Bain SC. Nitric oxide synthase gene polymorphisms and diabetic nephropathy. Diabetologia. 2003;46(3):426–428
- Ksiazek P, Wojewoda P, Muc K, Buraczynska M. Endothelial nitric oxide synthase gene intron 4 polymorphism in type 2 diabetes mellitus. Mol Diagn. 2003;7(2):119–123
- Frost D, Chitu J, Meyer M, Beischer W, Pfohl M. Endothelial nitric oxide synthase (ecNOS) 4 a/b gene polymorphism and carotid artery intima-media thickness in type-1 diabetic patients. Exp Clin Endocrinol Diabetes. 2003;111(1):12–15
- Degen B, Schmidt S, Ritz E. A polymorphism in the gene for the endothelial nitric oxide synthase and diabetic nephropathy. Nephrol Dial Transplant. 2001;16(1):185
- Rahimi Z, Rahimi Z, Shahvaisi-Zadeh F, Sadeghei S, Vessal M, Yavari N. eNOS 4a/b polymorphism and its interaction with eNOS G894T variants in type 2 diabetes mellitus: Modifying the risk of diabetic nephropathy. Disease Markers. 2013;34(6):437–443
- Lamnissou K, Zirogiannis P, Trygonis S, et al. Evidence for association of endothelial cell nitric oxide synthase gene polymorphism with earlier progression to end-stage renal disease in a cohort of Hellens from Greece and Cyprus. Genet Test. 2004;8(3):319–324
- Bellini MH, Figueira MN, Piccoli MF, et al. Association of endothelial nitric oxide synthase gene intron 4 polymorphism with end-stage renal disease. Nephrology (Carlton). 2007;12(3):289–293
- Tsukada T, Yokoyama K, Arai T, et al. Evidence of association of the ecNOS gene polymorphism with plasma NO metabolite levels in humans. Biochem Biophys Res Commun. 1998;245(1):190–193